[Literature Review] CRISPR Screening Innovation: A Novel Tool for Higher-Order Genetic Perturbations
Functional interactions among genetic elements are crucial for both natural and engineered phenotypes, often involving complex combinations of coding and non-coding sequences. However, testing higher-order genetic perturbations has faced limitations, especially in mammalian systems. Previous CRISPR-Cas9 screenings have been restricted to three perturbations per cell. Cas12a, with its ability to generate multiple CRISPR RNAs from a single promoter, offers potential for more extensive perturbations. However, its DNase activity can lead to genotoxicity.
To mitigate this, DNase-dead Cas fusion proteins, like dCas9-based CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa), have been used. A platform utilizing DNase-dead Cas12a (dCas12a) for multisite CRISPRi would be highly beneficial for studying combinatorial gene functions.
While dCas12a shows promise in individual assays, its effectiveness in pooled CRISPRi screenings remains uncertain and requires further investigation. Current dCas12a CRISPRi fusion constructs show suboptimal performance when delivered via lentivirus at limited doses, restricting their use in pooled settings.
Original Article Link: https://doi.org/10.1038/s41587-024-02224-0
To address this, researchers have engineered a variant of Acidaminococcus Cas12a (AsCas12a) with the R1226A mutation. This mutation enhances the stability of the ribonucleoprotein–DNA complex as a nicked DNA intermediate in vitro. Their findings were published in Nature Biotechnology under the title "Engineered CRISPR-Cas12a for Higher-Order Combinatorial Chromatin Perturbations". This combinatorial CRISPRi platform facilitates the efficient discovery of enhancer elements and enables the exploration of higher-order combinatorial perturbations of cis-regulatory elements, establishing a framework for effectively navigating large combinatorial spaces of chromatin perturbations.
I. Lentivirally Delivered CRISPRi by dAsCas12a Fusion Proteins is Hypoactive
In this study, researchers developed a CRISPRi functional genomics platform utilizing AsCas12a, the only Cas12a ortholog proven effective in pooled screens within mammalian cells. Previous research had employed dAsCas12a for CRISPRi via plasmid transient transfection in HEK 293T cells. However, when this construct was tested with lentivirally delivered crRNA in K562 cells, no changes in gene expression were observed.
Through a series of experiments, including lentiviral transduction, the researchers found that unlike plasmid transfection in HEK 293T cells, CRISPRi activity using dAsCas12a-3xKRAB with lentiviral crRNA requires distinct conditions for optimal performance.
To address the lack of activity, the researchers explored various mutations of Cas12a, eventually identifying denAsCas12a-KRAB as the most effective variant. This mutation exhibited strong repression of CD55, but results for CD81 were inconsistent.
The study further revealed that increasing the multiplicity of infection (MOI) for denAsCas12a-KRAB improved CRISPRi knockdown efficiency. However, at lower MOIs, which are essential for pooled screens, the activity diminished or was completely lost. To resolve these inconsistencies, the researchers also examined alternative methods to eliminate AsCas12a's DNase activity, including using truncated crRNA spacers. Unfortunately, these approaches produced weak and inconsistent CRISPRi activity across different cell lines.
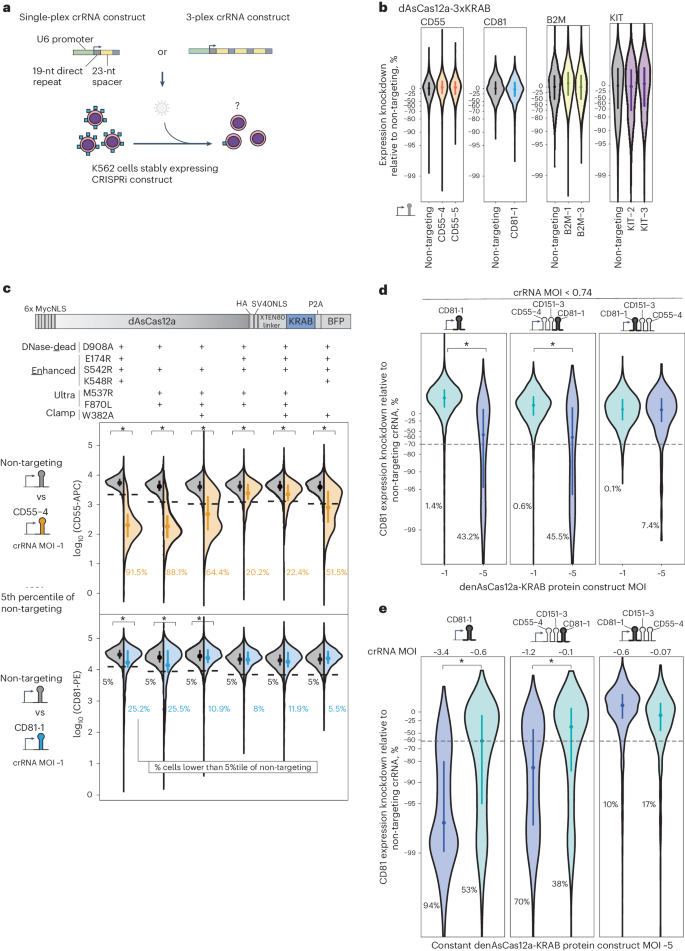
Fig.1: dAsCas12a-KRAB variants show weak and dose-limited CRISPRi activity with lentiviral crRNA delivery
II. MultiAsCas12a-KRAB (R1226A/E174R/S542R/K548R) Substantially Improves Lentivirally Delivered CRISPRi
To confirm that full activation of DNA cleavage in dAsCas12a can hinder strong CRISPRi activity by reducing DNA affinity, researchers conducted in vitro studies. They found that the dCas12a-DNA complex is 20 times more stable when the non-target strand is pre-cleaved.
The R1226A mutation, known for its significantly reduced cleavage activity, was incorporated into the Cas12a variant. The researchers discovered that this mutation enhances DNA binding compared to the fully inactive D908A variant. These findings suggest that the R1226A mutation could extend chromatin occupancy and improve the recruitment of transcriptional repressive complexes via the KRAB domain.
In experiments comparing the multiAsCas12a (multiplexed transcriptional interference AsCas12a) variant to denAsCas12a-KRAB, the multiAsCas12a variant demonstrated consistent and robust CRISPRi activity across various constructs. It showed less sensitivity to low levels of protein and crRNA, with minimal off-target effects. This modification not only revived the efficacy of several previously ineffective crRNA constructs, but also maintained low indel frequencies at target sites, indicating effective gene knockdown with minimal disruption to gene expression.
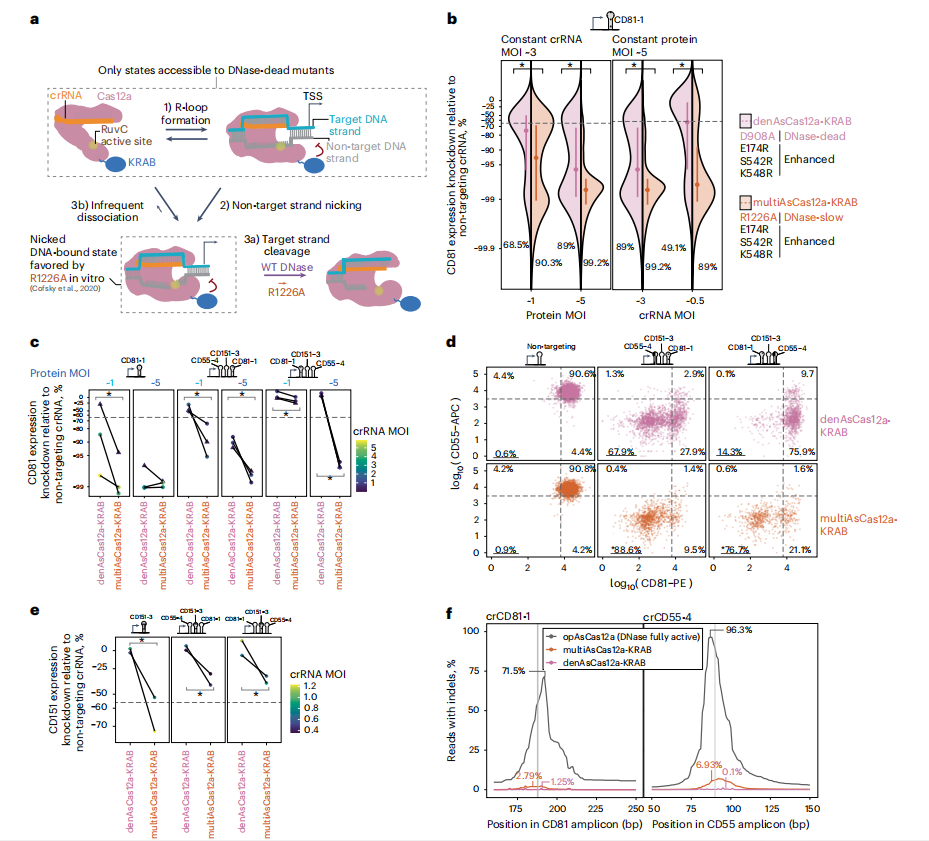
Fig. 2: MultiAsCas12a-KRAB (R1226A/E174R/S542R/K548R) significantly enhances lentivirally delivered CRISPRi by promoting a nicked DNA intermediate
III. MultiAsCas12a-KRAB Enables Multigene Transcriptional Repression Using Higher-Order Arrayed crRNA Lentiviral Constructs
To evaluate the activity of multiAsCas12a-KRAB in CRISPRi, the researchers used lentiviral crRNA arrays to test its performance in targeting three or more genomic sites per cell. The results showed that multiAsCas12a-KRAB demonstrated significantly superior CRISPRi activity across various constructs, with a strong reliance on the KRAB domain for effective gene knockdown.
Indel analysis indicated that the gene knockdown observed was primarily due to nongenetic perturbation of transcription, rather than residual DNA cleavage. At the single-cell level, multiAsCas12a-KRAB outperformed denAsCas12a-KRAB, achieving successful knockdowns at multiple targets, particularly in higher-order constructs, although some constructs exhibited reduced efficacy. These findings suggest that multiAsCas12a-KRAB holds promise for multiplexed CRISPRi applications, while also highlighting the unpredictable effects of pre-crRNA sequence context on CRISPRi efficiency.
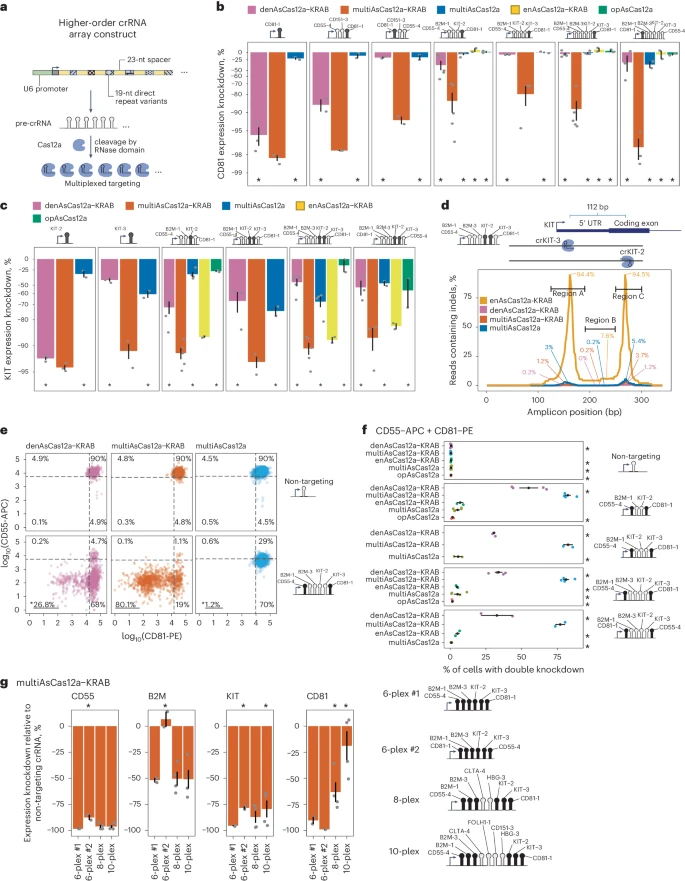
Fig. 3: MultiAsCas12a-KRAB enables multigene CRISPRi perturbations through higher-order crRNA lentiviral constructs
IV. MultiAsCas12a-KRAB Outperforms denAsCas12a-KRAB and Performs Similarly to dCas9-KRAB in Pooled Single-Guide CRISPRi Screens
The researchers evaluated the performance of multiAsCas12a-KRAB in high-throughput pooled screening. A library of 77,387 single crRNA lentiviral constructs was designed to assess Cas12a CRISPRi activity relative to the transcription start site (TSS) based on cell fitness. Pooled fitness screening in K562 cells expressing multiAsCas12a-KRAB demonstrated higher concordance in cell fitness scores compared to denAsCas12a-KRAB, indicating better performance and lower nonspecific genotoxicity.
The results showed that multiAsCas12a-KRAB effectively generated a metagene profile of CRISPRi activity around the TSS, similar to that of dCas9-KRAB, and outperformed denAsCas12a-KRAB. Although a smaller proportion of crRNAs targeting non-canonical PAMs exhibited activity, individual crRNA efficacy varied significantly within PAM sequences, which could not be fully predicted by CRISPick models.
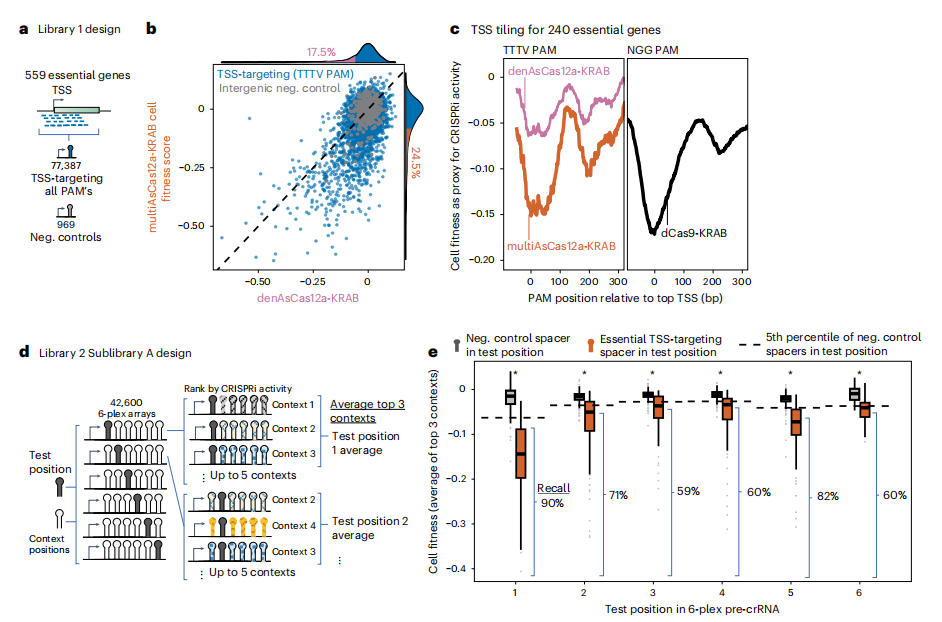
Fig. 4: MultiAsCas12a-KRAB enables TSS-targeting pooled CRISPRi screens, including 6-plex crRNA arrays
V. MultiAsCas12a-KRAB Enables Pooled CRISPRi Screens Using 6-plex crRNA Arrays
To evaluate the performance of multiAsCas12a-KRAB in pooled CRISPRi screens, a new library was constructed and screened in K562 cells engineered to express multiAsCas12a-KRAB. The variant demonstrated high replicate concordance and no significant nonspecific genotoxicity. The results showed that test position spacers had weaker cell fitness defects compared to their single crRNA counterparts, with a recall rate of 59% to 90% for active single crRNAs. This suggests that most individually active crRNAs retained measurable efficacy when integrated into 6-plex arrays in pooled screens.
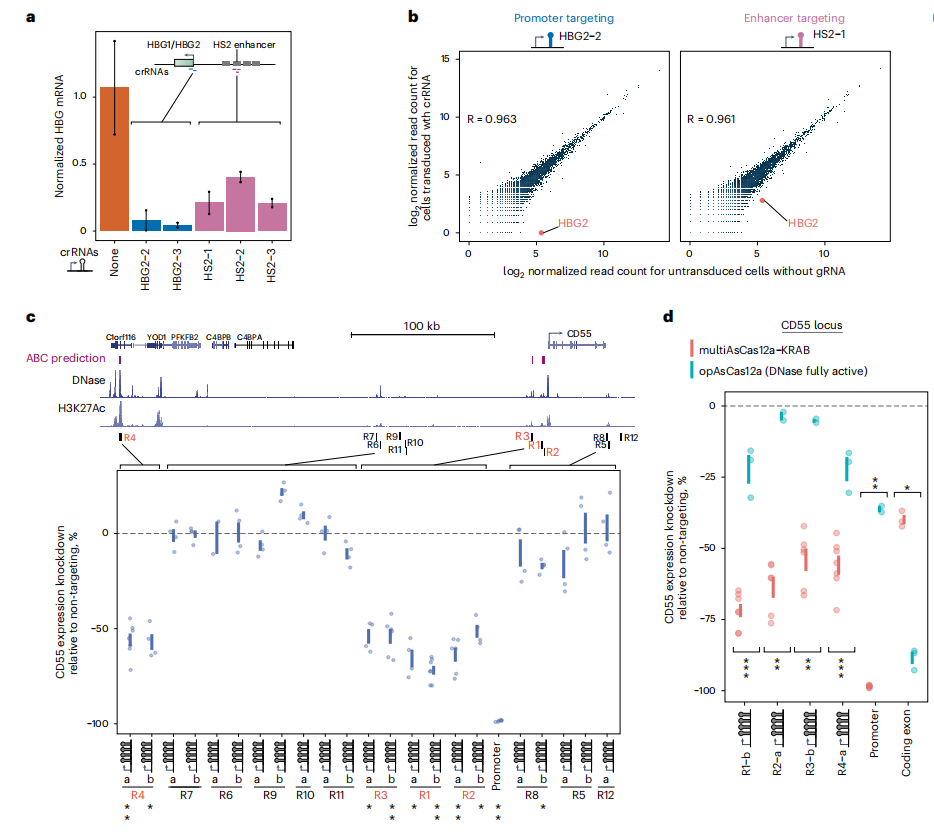
Fig. 5: MultiAsCas12a-KRAB CRISPRi enables enhancer perturbation and discovery
VI. Discovery and Higher-Order Combinatorial Perturbations of Cis-Regulatory Elements
The human genome is estimated to contain approximately 500,000 enhancers, yet only a fraction have been functionally tested. This study confirmed that multiAsCas12a-KRAB can perturb both gene promoters and enhancers through CRISPRi, specifically demonstrating its ability to target the HBG1/HBG2 paralogs and the HS2 enhancer. Using the CD55 locus in K562 cells, the researchers identified several previously uncharacterized enhancers that regulate CD55 expression, marking the first functionally validated enhancers in myeloid cells.
Furthermore, they employed multiAsCas12a-KRAB to explore the effects of higher-order perturbations on the MYC locus, revealing that co-targeting multiple cis-regulatory elements led to stronger gene knockdown than targeting the promoter alone.
These findings underscored the utility of multiAsCas12a-KRAB in enhancer studies and suggested that combinatorial targeting of regulatory elements can more effectively modulate gene expression, establishing a framework for efficient testing of genetic perturbations.
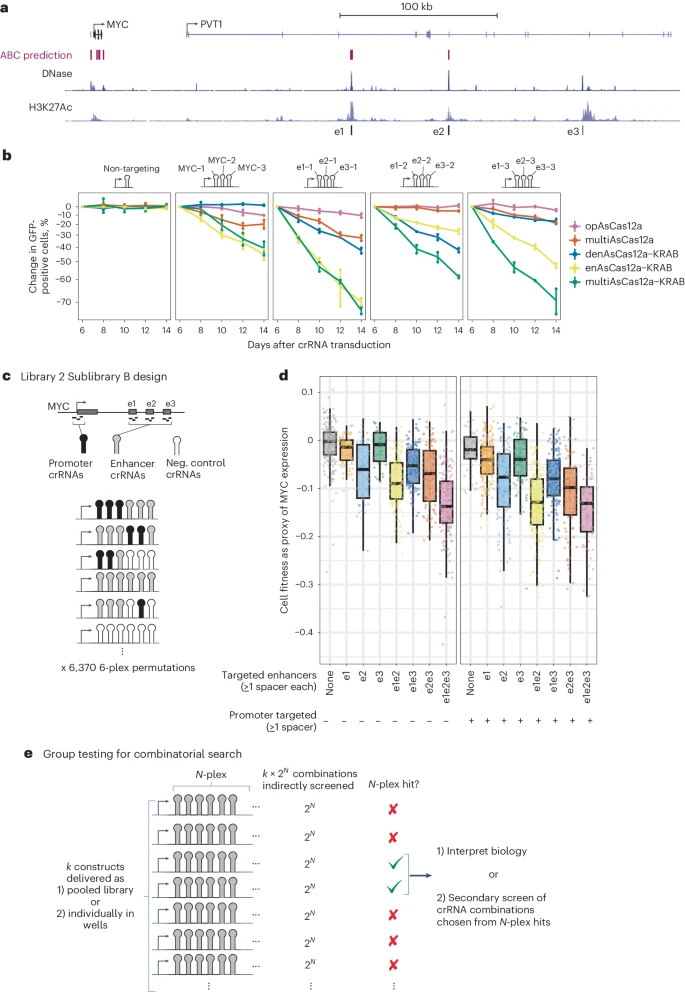
Fig. 6: Higher-order combinatorial targeting of cis-regulatory elements by multiAsCas12a-KRAB establishes a group testing framework
Overall, research into CRISPR-Cas-mediated chromatin perturbations has rapidly advanced. By flexibly combining various effector domains, multiAsCas12a enables group testing for many chromatin perturbation objectives. Together with the group testing framework, multiAsCas12a facilitates the engineering and understanding of combinatorial genetic processes across a broad range of biological fields, at scales that were previously difficult to achieve.
This study established multiAsCas12a-KRAB as a powerful platform for higher-order combinatorial CRISPRi perturbations, demonstrating its superior efficacy in regulating gene transcription and enhancer function, making it a reliable tool for precise genetic modulation.
EDITGENE offers a complete solution for CRISPR library screening, CRISPR KO/CRISPRa/CRISPRi, and the most extensive collection of library plasmids/viruses, which is available within weeks.
See how you can utilize CRISPR library screening technology in your research for target genes and pathways. Check out EDITGENE's complete solution for CRISPR library screening, CRISPR KO/CRISPRa/CRISPRi, and the most extensive collection of library plasmids/viruses, which is available within weeks.
Recent Blogs:
3.[Literature Review] CRISPR Knockout Provides Novel Insight into CENP-E-mediated Cell Division
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com










![[Literature Review] A “Gene Scalpel” Emerges: A New Hope for Tay-Sachs Disease Cure](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)