 CRISPR Detection Service
CRISPR Detection Service
Service Details
| Service Content | Service Description | Delivery Standard |
|---|---|---|
| Preparation of Target Gene Template | Synthesis of target plasmid | Target plasmid |
| Design and Synthesis of crRNA and RPA Isothermal Amplification Primers | Design of crRNA and RPA primers based on the target sequence | crRNA,2 OD |
| Activity Screening of RPA Isothermal Amplification Primers | Multiple RPA primers are used to amplify the target sequence, and the most efficient primer sequence is selected | RPA isothermal amplification primers, 2 OD |
| Activity Screening of crRNA | Multiple crRNAs are analyzed for activity, and the most efficient crRNA is identified | Final report and raw data |
| Establishment and Optimization of Experimental System for crRNA and RPA Isothermal Amplification Primers | Sensitivity, specificity, and accuracy tests for crRNA and RPA | Final report and raw data |
| FASST Full-suite Detection Services | / | Final report and CRISPR reagent kit for 50 reactions |
FASST Rapid Detection Platform

EDI-Service Advantages
Fast
Sensitive
Simple
Accurate
Technology Comparison
| Item | FASST | PCR | LAMP (Isothermal Amplification) | Traditional CRISPR Detection |
|---|---|---|---|---|
|
Reaction Temperature |
37-42℃ (Isothermal) |
95℃-55℃-72℃ (Variable) |
65℃ (Isothermal) |
37-42℃ (Isothermal) |
| Run Time | 5-20 mins | 1-2h | 40-60mins | 30-60 mins |
|
Primer Quantity |
2 primers |
2 primers |
4-6 primers |
2 primers |
| Reagent Form | Liquid/ Lyophilized | Liquid | Liquid | Liquid/Lyophilized |
| Equipment Requirement | Isothermal equipment (metal bath, water bath, etc.) | PCR Amplifier | Isothermal equipment | Isothermal equipment (metal bath, water bath, etc.) |
| Ease of Use | Extremely simple, truly portable | Requires professional operation, complex equipment | Simple to operate | Extremely simple, truly portable |
| Aerosol Contamination | Single-tube reaction, low contamination risk | Risk of aerosol contamination | Risk of aerosol contamination | Two-tube reaction, risk of aerosol contamination |
Workflow
Case Study
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Selected Customer Resources
Deep whole-genome analysis of 494 hepatocellular carcinomas
Abstract:
To date, more than half of global hepatocellular carcinoma (HCC) cases occur in China, yet comprehensive whole-genome analyses focusing on HBV-related HCC within the Chinese population remain scarce. To address this challenge, researchers initiated the China Liver Cancer Atlas (CLCA) project, aiming to conduct large-scale whole-genome sequencing to unravel the unique pathogenic mechanisms and evolutionary trajectories of HCC in China.
The researchers performed deep whole-genome sequencing on 494 HCC tumor samples, with an average depth of 120×, alongside matched blood controls, providing a detailed genomic landscape of HBV-associated HCC. Beyond confirming well-known coding driver genes such as TP53 and CTNNB1, the study identified six novel coding drivers—including FGA—and 31 non-coding driver genes.
Additionally, the research uncovered five new mutational signatures, including SBS_H8, and characterized the presence of extrachromosomal circular DNA (ecDNA) formed via HBV integration, which contributes to oncogene amplification and overexpression. Functional validation experiments demonstrated that mutations in genes such as FGA, PPP1R12B, and KCNJ12 significantly enhance HCC cell proliferation, migration, and invasion.
These findings not only deepen our insights into the genomics of HCC, but also open up new potential targets for diagnosis and therapy. View details>>
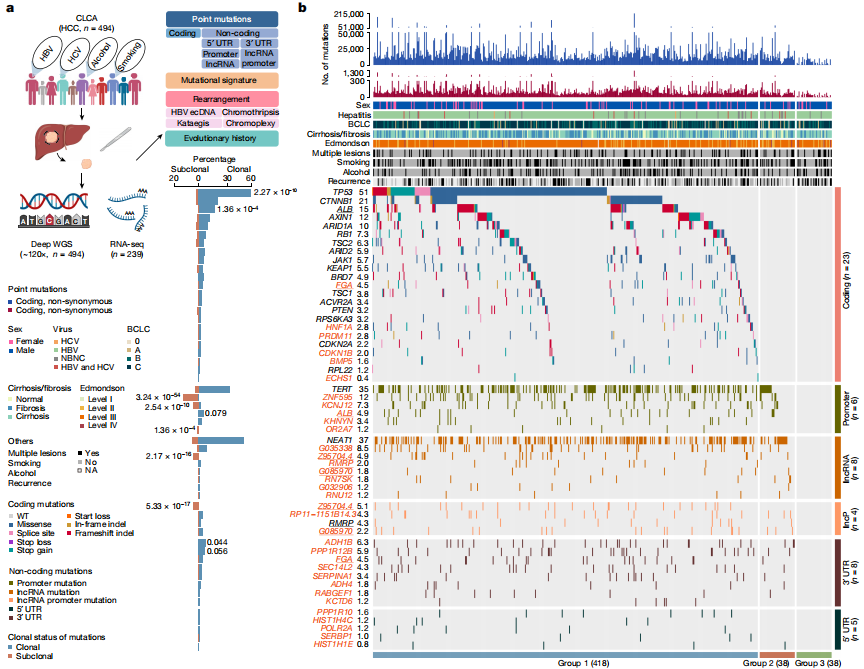
Candidate driver landscape
Targeted Macrophage CRISPR-Cas13 mRNA Editing in Immunotherapy for Tendon Injury
Abstract:
During the acute inflammatory phase of tendon injury, excessive activation of macrophages leads to the overexpression of SPP1, which encodes osteopontin (OPN), thereby impairing tissue regeneration. The CRISPR-Cas13 system holds great promise for tissue repair due to its unique RNA editing and rapid degradation capabilities; however, its application has been limited by the lack of efficient delivery methods.
To address this, the researchers systematically screened various cationic polymers targeting macrophages and developed a nanocluster carrier capable of efficiently delivering Cas13 ribonucleoprotein complexes (Cas13 RNPs) into macrophages. Utilizing a reactive oxygen species (ROS)-responsive release mechanism, this system specifically suppresses the overexpression of SPP1 in macrophages within the acute inflammatory microenvironment of tendon injury.
Experimental results demonstrated that this targeted delivery strategy significantly reduced the population of SPP1-overexpressing macrophages induced by injury, inhibited fibroblast activation, and alleviated peritendinous adhesion formation. Furthermore, the study elucidated that SPP1 promotes fibroblast activation and migration through the CD44/AKT signaling pathway, and that inhibiting this pathway effectively mitigates adhesion formation following tendon injury. View details>>
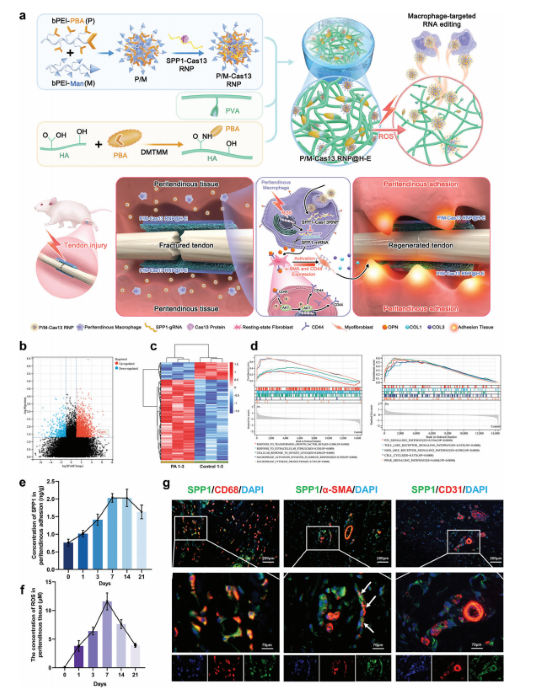
Schematic diagram illustrating immune microenvironment-activated mRNA editing strategies of macrophages for PA therapy
Electrical stimulation of piezoelectric BaTiO3 coated Ti6Al4V scaffolds promotes anti-inflammatory polarization of macrophage and bone repair via MAPK/JNK inhibition and OXPHOS activation
Abstract:
Spinal cord injury (SCI) is a severe disabling condition that causes permanent loss of sensory, autonomic, and motor functions. While stem cell therapies, particularly mesenchymal stem cells (MSCs), show great promise for SCI treatment, their limited regenerative capacity restricts their application in tissue repair. The researchers observed that extracellular vesicles derived from antler bud progenitor cells (EVsABPC) may carry bioactive signals that promote tissue regeneration. Accordingly, they isolated and engineered EVs from ABPCs for SCI therapeutic investigation.
The study found that EVsABPC significantly enhanced neural stem cell (NSC) proliferation, promoted axonal growth, reduced neuronal apoptosis, and modulated inflammation by shifting macrophage polarization from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype. Moreover, engineered EVsABPC modified with cell-penetrating peptides demonstrated improved targeting to the SCI lesion site, markedly enhancing neural regeneration and functional motor recovery. These findings highlight EVsABPC as a promising candidate for SCI therapy. View details>>
Graphical abstract
Generation of recombinant antibodies by mammalian expression system for detecting S-metolachlor in environmental waters
Abstract:
S-metolachlor (S-MET) is one of the most widely produced and applied herbicides in China. Owing to its chemical properties, it tends to persist in soil and easily contaminates surface and groundwater through leaching and runoff. This environmental persistence poses a serious threat to plant development and, through the food chain, to human health.
To address the limitations of current detection technologies and meet the growing demand for high-efficiency analytical tools, the researchers employed a mammalian expression system to generate recombinant antibodies targeting S-MET.
Building on the successful expression of these antibodies, they established a sensitive immunoassay for monitoring S-MET residues in various environmental water samples. The icELISA results showed that the recombinant antibodies retained the sensitivity, specificity, and biological activity of the original monoclonal antibodies, delivering accurate and reproducible detection in river water, agricultural runoff, and tap water. View details>>
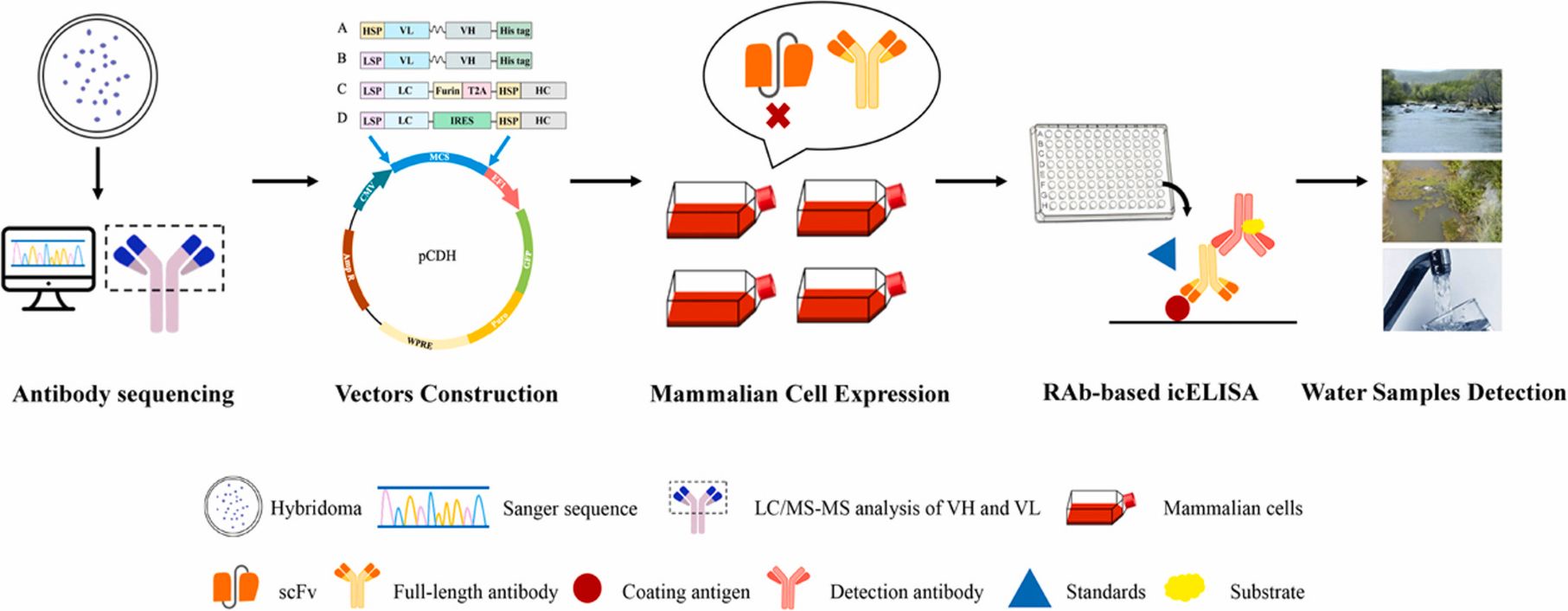
Graphical abstract
Dumbbell probe initiated multi-rolling circle amplification assisted CRISPR/Cas12a for highly sensitive detection of clinical microRNA
Abstract:
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that regulate gene expression by interacting with the mRNAs of target genes. Given their crucial role in the development and progression of various diseases, miRNAs have emerged as promising biomarkers for clinical diagnostics.
In this study, researchers established a novel detection platform, termed DBmRCA, which combines dumbbell probe-initiated multi-rolling circle amplification with the high-sensitivity signal output of CRISPR/Cas12a. This enzyme-free, isothermal method enables accurate quantification of miRNA within just 30 minutes.
Clinical validation revealed that the expression levels of miR-200a and miR-126 were significantly downregulated in lung cancer tissues, and results from DBmRCA were consistent with those obtained by conventional techniques. With its high sensitivity, rapid turnaround, and simplified workflow, the DBmRCA platform presents a reliable tool for miRNA detection and holds strong promise for early diagnosis and therapeutic monitoring of lung cancer. View details>>
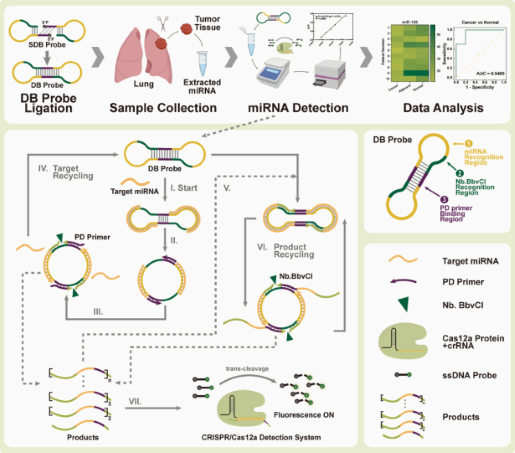
Graphical abstract
FAQ
How to Improve the Detection Sensitivity of Cas Enzymes?
2.Choose an appropriate signal reporter substrate. Research indicates that using a 15 nt single-stranded DNA (ssDNA) as a reporter substrate maximizes the cleavage reaction rate of Cas12a, significantly enhancing the reaction rate compared to the commonly used 5-nt ssDNA.
3.Optimize reaction conditions and buffers. Adjusting the CRISPR reaction parameters, such as the ratio of Cas enzyme to crRNA, the concentration of the Cas enzyme, and the reaction temperature, can improve detection performance to some extent.
How to Design crRNA?
1.Identify the target gene sequence.
2.Specify the Cas protein being used. Different Cas proteins require corresponding PAM (Protospacer Adjacent Motif) sequences; for instance, Cas12a needs the "TTTV" PAM sequence for target recognition.
3.Select the crRNA targeting region. Choose a 20 nt nucleotide sequence on the target gene that is adjacent to the PAM site and pairs with the complementary strand of the crRNA.
4.Combine the selected 20 nt target sequence (variable part) with the scaffold sequence (fixed part) to design the crRNA sequence.
5.Use online tools such as CRISPR design tools (e.g., CRISPOR, Benchling, etc.) to assist in designing crRNA. These tools can predict the efficiency and specificity of the sgRNA, helping to avoid potential off-target effects.
6.After completing the design, the synthetic crRNA sequence can be ordered from a synthetic biology company.




