 Overexpression Cell Line
Overexpression Cell Line
In-Stock stable cell lines
Service Details
| Cell Types | Various cell types, including tumor, conventional, stem, primary, and immortalized cell lines. |
|---|---|
| Services | ORF stable cell line/CRISPRa stable cell line |
| Deliverables |
Gene overexpression polyclonal cell pool (2 vials per pool, 1×10^6 cells/vial) Gene overexpression monoclonal cell line ≥1 clone (2 vials/clone, 1×10^6 cells/vial) Experimental report |
| Turnaround/Price |
As fast as 4 weeks,as low as $2000 |
EDI-Service Advantages
Versatile Vector Selection
Efficient Transfection
Precise Clone Screening
Experienced Team
Service Types
An ORF stable cell line enables long-term expression of a target gene of up to 10 kb in size within cells.
An ORF overexpression cell line is created by stably integrating a selected transcript of an exogenous target gene into the host cell genome, ensuring its continuous and efficient expression. Such cell lines can maintain stable target gene expression over extended passaging (typically more than 30 generations) or during drug selection, and can be established using either a lentiviral system or a transposon-based system. The core scientific value of stable overexpression cell lines lies in the genomic integration of an exogenous gene, which overcomes the intrinsic regulatory constraints of the host cell to achieve long-term, stable, and exceptionally high levels of gene expression. Despite potential challenges such as insertional mutagenesis and transcriptional silencing, they remain the gold-standard tools for gene function studies, disease model development, and industrial-scale protein production. Contact Us
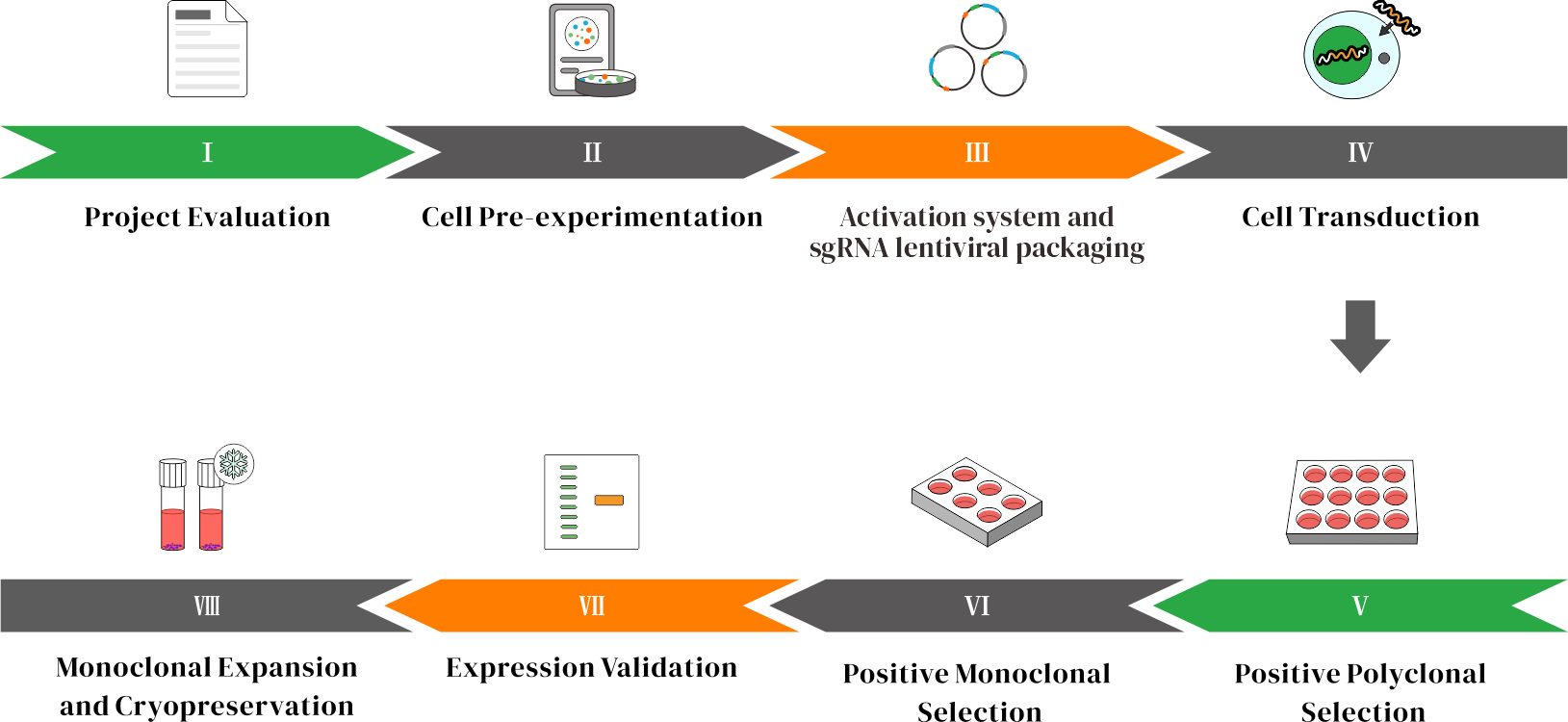
Workflow
A CRISPRa stable cell line enables long-term expression of target genes larger than 10 kb within cells.
CRISPRa (CRISPR activation) is a gene regulation technology based on the CRISPR system. It employs an engineered, catalytically inactive Cas9 protein (dCas9) in combination with a single guide RNA (sgRNA) to target and bind genomic regions near the transcription start site of the target gene. By fusing dCas9 with transcriptional activators (such as VP64, p65, and Rta) or synthetic activation systems like SAM, CRISPRa induces overexpression of the cell’s own endogenous genes. With carefully designed sgRNAs, CRISPRa can simultaneously target multiple gene loci, enabling coordinated regulation of complex gene networks and providing a powerful tool for functional genomics research. The activation system and sgRNAs can be delivered via lentiviral packaging for stable and efficient expression.
Workflow
Case Study
Lentivirus-Mediated Overexpression of copEGFP in A375 Cells
The copEGFP gene sequence was cloned into the lentiviral expression vector pLV3-CAG-MCS-P2A-Puro, followed by packaging and production of high-titer lentiviral particles. These particles were used to infect the human melanoma cell line A375. Expression of the EGFP reporter gene in infected cells was directly observed using fluorescence microscopy to evaluate overexpression efficiency.
Note: TheA375-copEGFP cellsused in this case are ready-to-use stocks provided by EDITGENE (Catalog No.: EDC01053), ensuring rapid experimental setup and reproducibility.copGFP Overexpression Cell Line Stock Library
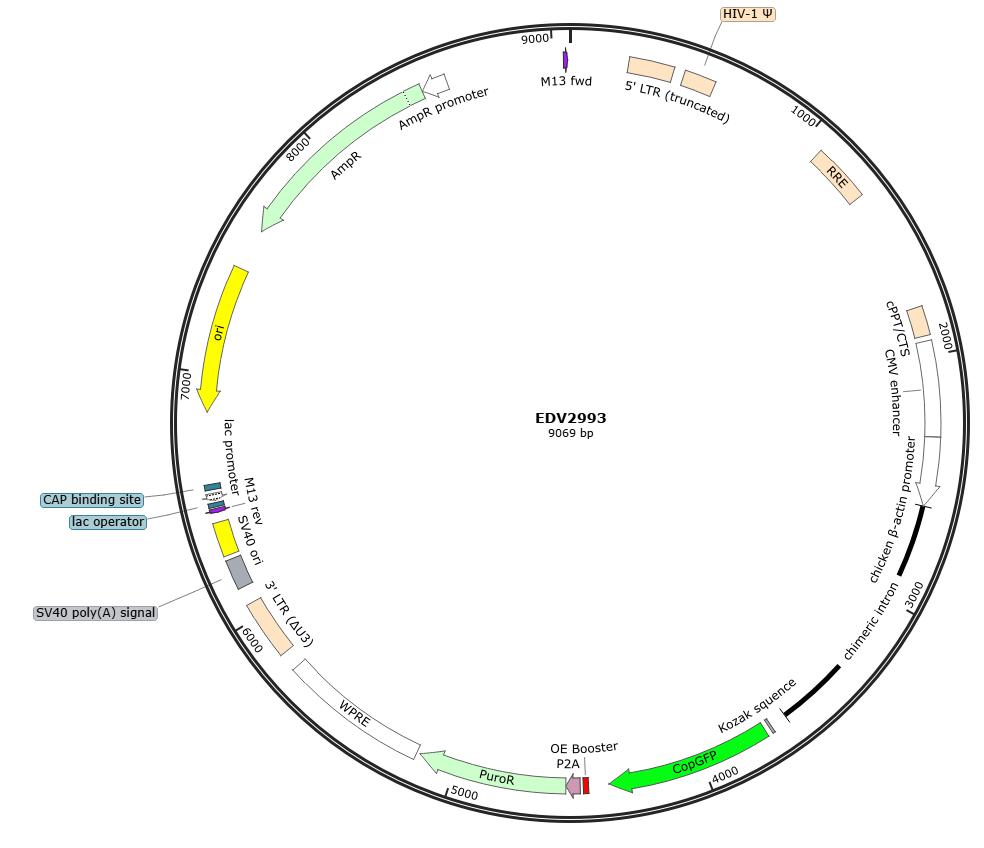
Figure 1: Map of the pLV3-CAG-MCS-P2A-Puro backbone vector
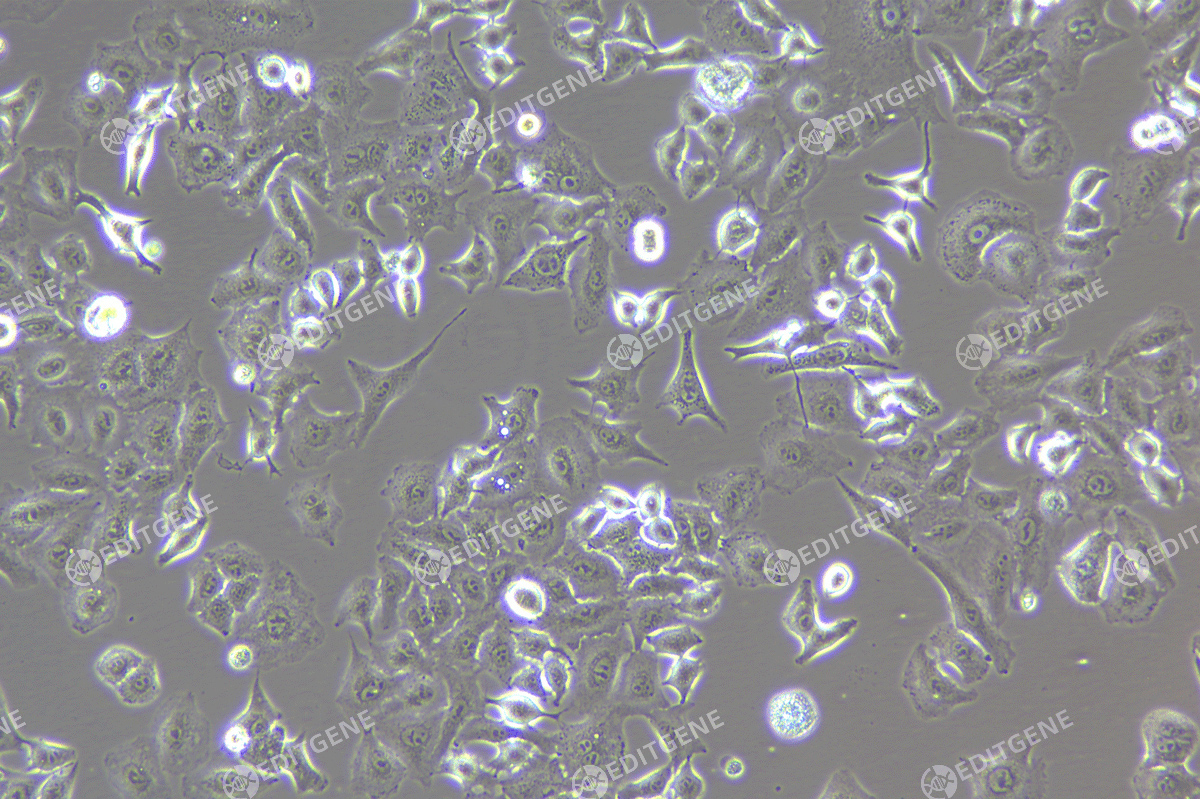
Figure 2: Bright-field image of A375 cells
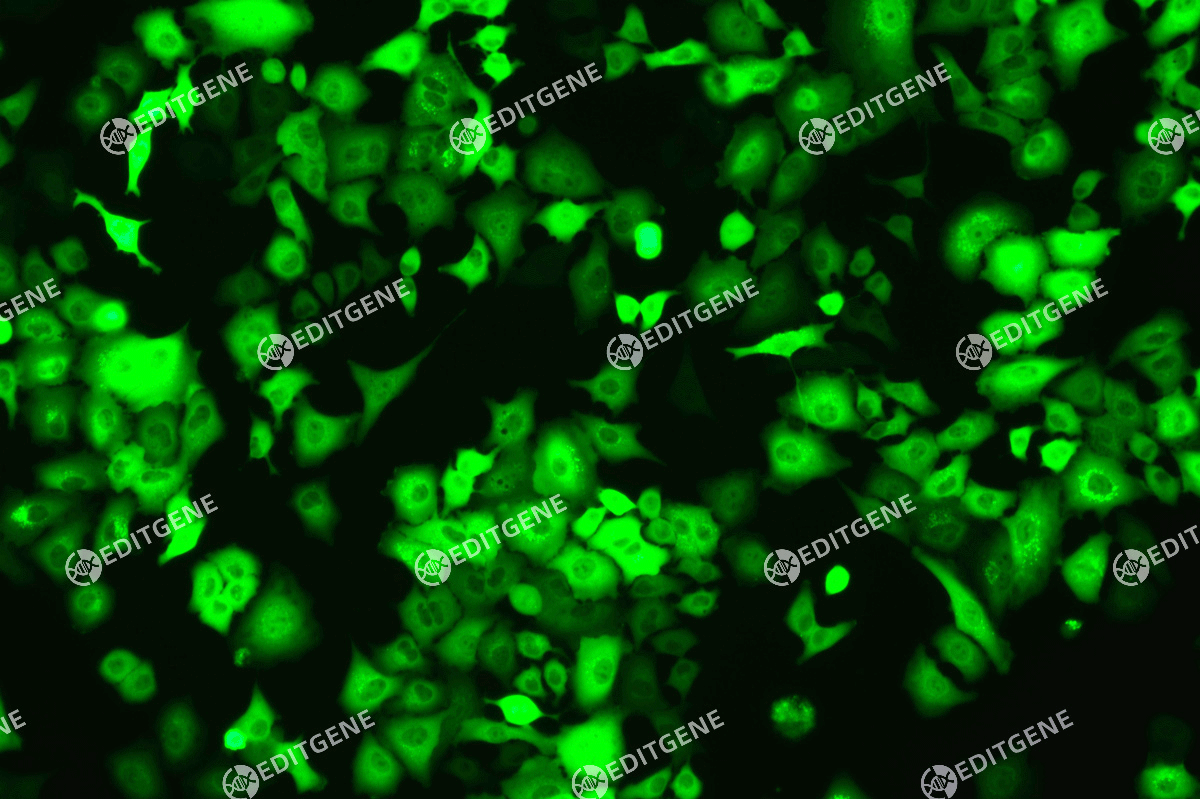
Figure 3: Fluorescence image of A375-copEGFP cells
Lentivirus-Mediated Overexpression of SCN10A in HEK293T Cells and Detection of Flag Tag
The coding sequence of the target gene SCN10A was cloned into the lentiviral expression vector pLV-EF1a-MCS-3×flag-puro to construct an expression vector with a C-terminal 3×Flag tag fusion. After packaging high-titer lentiviral particles, the virus was used to infect human embryonic kidney HEK293T cells. Expression of the fusion protein was validated by Western blot (WB) using an anti-Flag antibody to assess the overexpression efficiency of the target gene.
Note: The HEK293T-SCN10A cellsused in this study are ready-to-use stocks provided by EDITGENE (Catalog No.: EDC01474), ensuring rapid experimental progress and reproducibility.Overexpression Cell Line Stock Library
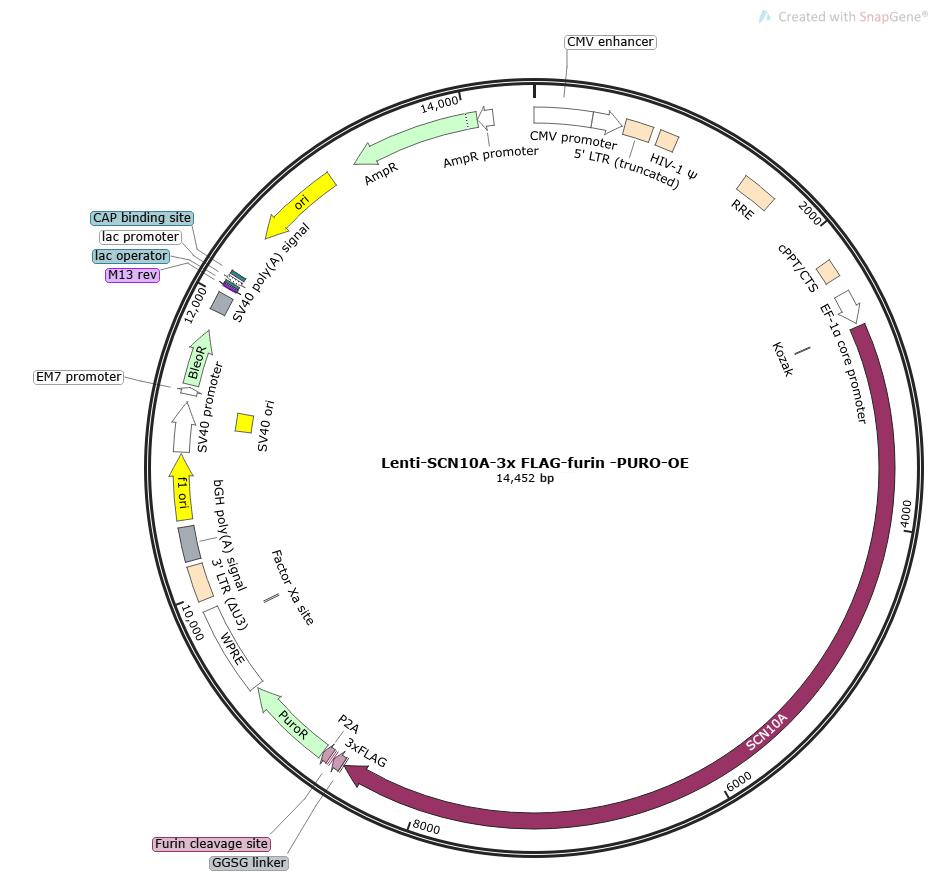
Figure 1: Map of the pLV-EF1a-MCS-3×flag-puro backbone vector

Figure 2: qRT-PCR validation results
LRP1 Gene Overexpression Mediated by CRISPRa System
The lentivirus-mediated CRISPR activation (CRISPRa) system was employed to specifically enhance the transcription of the LRP1 gene.
①1.Core Effector Delivery: Lentiviral vectors EDV1951-lenti (expressing dCas9-VP64_Blast) and EDV2939-lentiMPH v2 (expressing the MS2-P65-HSF1 fusion protein v2) were packaged into viral particles and co-infected into the target cell line to establish stable cell lines expressing the dCas9-VP64 and MPH v2 effector complex. Screening was performed using Blasticidin and Hygromycin.
②2.Targeted sgRNA Delivery and Selection: Three distinct single guide RNAs (sgRNAs) targeting the promoter/regulatory regions of the LRP1 gene were designed. Each sgRNA sequence was cloned individually into the lentiviral sgRNA expression vector EDV3689-Lenti sgRNA(MS2)-Puro to generate LRP1-targeting sgRNA constructs. Lentiviral particles were produced for each sgRNA and used to infect the stable dCas9-VP64/MPH v2-expressing cells. Puromycin selection was applied to obtain cell lines with stable integration of different sgRNAs.
③3.Detection: Changes in LRP1 gene expression at both mRNA and protein levels were assessed by qRT-PCR. The results confirmed effective activation of LRP1 gene expression in the target cells by this system.Overexpression Cell Line Stock Library
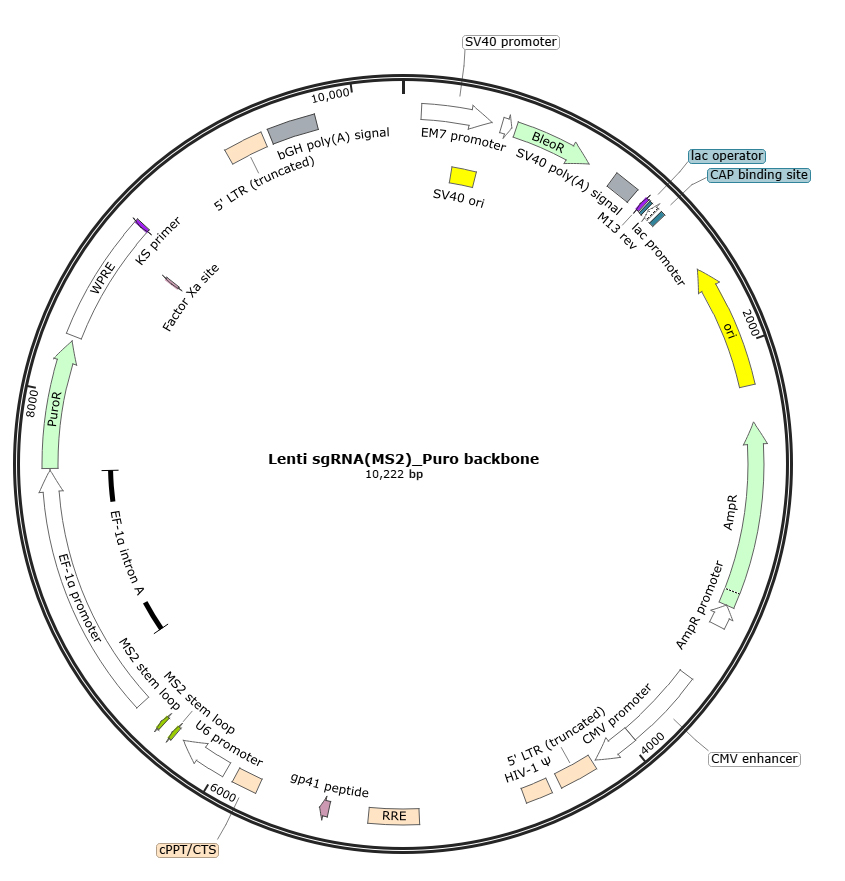
Figure 1: Map of the EDV3689-Lenti sgRNA(MS2)-Puro backbone vector
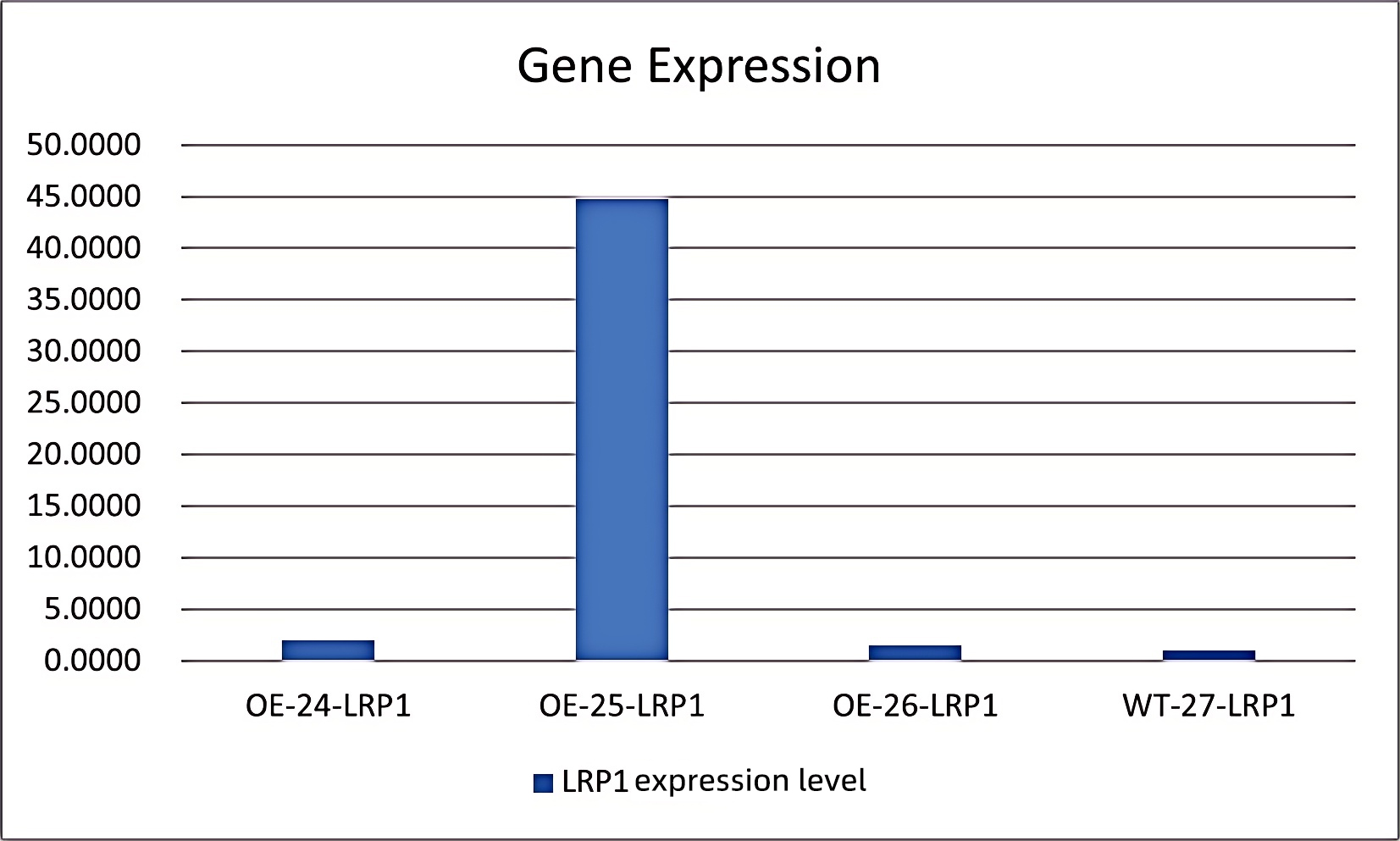
Figure 2: OE-24 (LRP1 transcriptionally activated stable line with gRNA1), OE-25 (with gRNA2), OE-26 (with gRNA3), and WT-27 (wild-type cells)
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Reference Materials
Overexpression or knockdown of VASN gene in rectal cancer
This study examines the role of VASN in rectal cancer, finding that its high expression is associated with pulmonary metastasis and chemotherapy resistance. By establishing stable overexpression and knockdown cell lines of VASN, the authors further validated VASN's role in promoting cancer cell migration, invasion, and proliferation. Both in vitro and in vivo results show that VASN overexpression significantly enhances cancer cell invasion and reduces sensitivity to chemotherapeutic agents. The experimental methods include transfection of the VASN gene using lentiviral vectors, establishment of stable cell lines through G418 resistance selection, and verification of VASN overexpression via Western blot and qPCR.
LBX1 gene overexpression in mice
The LBX1 gene, located near single nucleotide polymorphisms highly associated with adolescent idiopathic scoliosis susceptibility, is considered one of the strongest candidate genes for the disease mechanism. The study found that LBX1 deletion not only causes spinal deformities but also affects lean body mass, suggesting LBX1's potential role in energy metabolism. This study aimed to test this hypothesis by analyzing the phenotype of LBX1-deficient skeletal muscle in mice, focusing on energy metabolism. Results showed that LBX1-deficient mice exhibited greater resistance to high-fat diet-induced obesity, despite similar food intake to control mice. Mutant mice also demonstrated better glucose tolerance, higher maximal aerobic capacity, and increased core temperature. Furthermore, LBX1 overexpression reduced glucose uptake in cultured cells. Together, the data indicate that LBX1 acts as a negative regulator of energy metabolism, with its deletion increasing systemic energy expenditure, leading to lean body mass. The study also suggests a potential association between LBX1 dysfunction and lean body mass in adolescent idiopathic scoliosis patients.
Overexpression or silencing of SRSF10 gene in SW480 and HCT116 cells
This study investigates the role and pathogenesis of serine- and arginine-rich splicing factor 10 (SRSF10) in colorectal cancer (CRC). Bioinformatics analysis predicted SRSF10 gene expression in CRC patients, and functional experiments involving SRSF10 knockdown and overexpression were conducted using CCK8, Transwell, scratch assays, and flow cytometry. SRSF10 knockdown inhibited CRC cell proliferation and migration, promoted apoptosis, and altered DNA replication, while SRSF10 overexpression enhanced proliferation and migration and caused cell cycle changes. Notably, high SRSF10 expression reduced glucose uptake in cultured cells, further indicating its role as a negative regulator of energy metabolism. The study also revealed changes in the RFC5 gene in CRC cells after SRSF10 knockdown, with SRSF10 increasing the exon 2-AS1(S) transcript variant of RFC5, promoting abnormal splicing of exon 2 and advancing CRC progression. These findings suggest that SRSF10 may serve as an important target for CRC clinical diagnosis and treatment.
Selected Customer Resources
Deep whole-genome analysis of 494 hepatocellular carcinomas
Abstract:
To date, more than half of global hepatocellular carcinoma (HCC) cases occur in China, yet comprehensive whole-genome analyses focusing on HBV-related HCC within the Chinese population remain scarce. To address this challenge, researchers initiated the China Liver Cancer Atlas (CLCA) project, aiming to conduct large-scale whole-genome sequencing to unravel the unique pathogenic mechanisms and evolutionary trajectories of HCC in China.
The researchers performed deep whole-genome sequencing on 494 HCC tumor samples, with an average depth of 120×, alongside matched blood controls, providing a detailed genomic landscape of HBV-associated HCC. Beyond confirming well-known coding driver genes such as TP53 and CTNNB1, the study identified six novel coding drivers—including FGA—and 31 non-coding driver genes.
Additionally, the research uncovered five new mutational signatures, including SBS_H8, and characterized the presence of extrachromosomal circular DNA (ecDNA) formed via HBV integration, which contributes to oncogene amplification and overexpression. Functional validation experiments demonstrated that mutations in genes such as FGA, PPP1R12B, and KCNJ12 significantly enhance HCC cell proliferation, migration, and invasion.
These findings not only deepen our insights into the genomics of HCC, but also open up new potential targets for diagnosis and therapy. View details>>
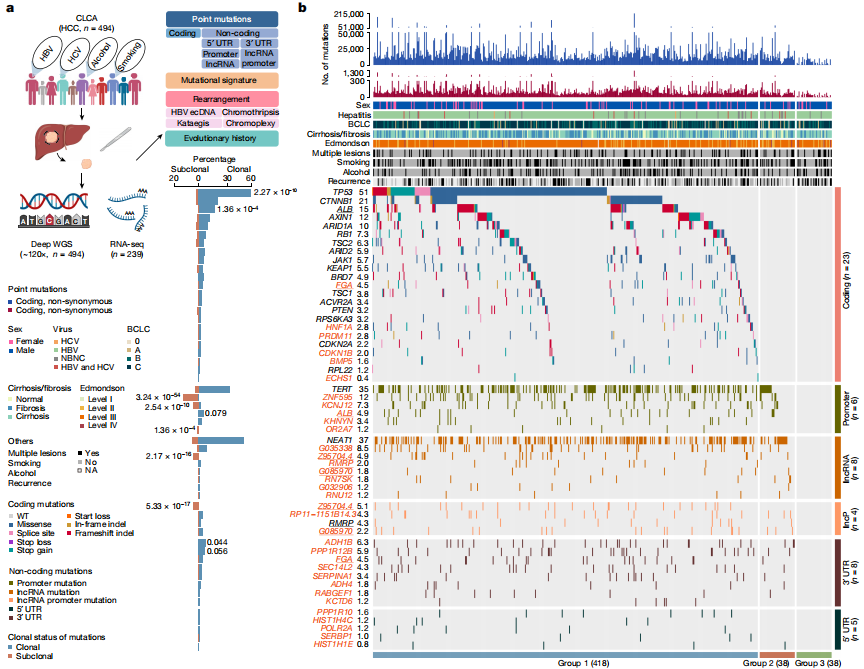
Candidate driver landscape
Targeted Macrophage CRISPR-Cas13 mRNA Editing in Immunotherapy for Tendon Injury
Abstract:
During the acute inflammatory phase of tendon injury, excessive activation of macrophages leads to the overexpression of SPP1, which encodes osteopontin (OPN), thereby impairing tissue regeneration. The CRISPR-Cas13 system holds great promise for tissue repair due to its unique RNA editing and rapid degradation capabilities; however, its application has been limited by the lack of efficient delivery methods.
To address this, the researchers systematically screened various cationic polymers targeting macrophages and developed a nanocluster carrier capable of efficiently delivering Cas13 ribonucleoprotein complexes (Cas13 RNPs) into macrophages. Utilizing a reactive oxygen species (ROS)-responsive release mechanism, this system specifically suppresses the overexpression of SPP1 in macrophages within the acute inflammatory microenvironment of tendon injury.
Experimental results demonstrated that this targeted delivery strategy significantly reduced the population of SPP1-overexpressing macrophages induced by injury, inhibited fibroblast activation, and alleviated peritendinous adhesion formation. Furthermore, the study elucidated that SPP1 promotes fibroblast activation and migration through the CD44/AKT signaling pathway, and that inhibiting this pathway effectively mitigates adhesion formation following tendon injury. View details>>
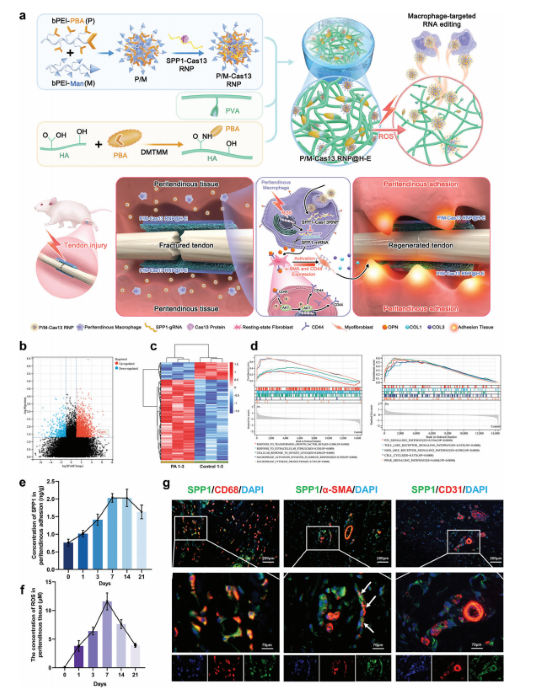
Schematic diagram illustrating immune microenvironment-activated mRNA editing strategies of macrophages for PA therapy
Electrical stimulation of piezoelectric BaTiO3 coated Ti6Al4V scaffolds promotes anti-inflammatory polarization of macrophage and bone repair via MAPK/JNK inhibition and OXPHOS activation
Abstract:
Spinal cord injury (SCI) is a severe disabling condition that causes permanent loss of sensory, autonomic, and motor functions. While stem cell therapies, particularly mesenchymal stem cells (MSCs), show great promise for SCI treatment, their limited regenerative capacity restricts their application in tissue repair. The researchers observed that extracellular vesicles derived from antler bud progenitor cells (EVsABPC) may carry bioactive signals that promote tissue regeneration. Accordingly, they isolated and engineered EVs from ABPCs for SCI therapeutic investigation.
The study found that EVsABPC significantly enhanced neural stem cell (NSC) proliferation, promoted axonal growth, reduced neuronal apoptosis, and modulated inflammation by shifting macrophage polarization from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype. Moreover, engineered EVsABPC modified with cell-penetrating peptides demonstrated improved targeting to the SCI lesion site, markedly enhancing neural regeneration and functional motor recovery. These findings highlight EVsABPC as a promising candidate for SCI therapy. View details>>
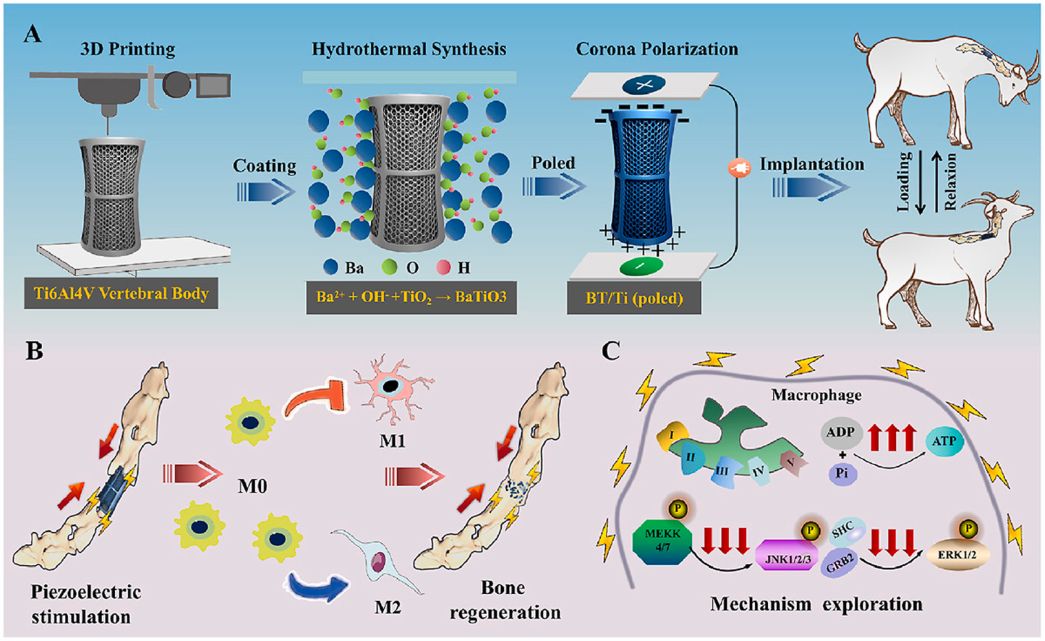
Graphical abstract
Generation of recombinant antibodies by mammalian expression system for detecting S-metolachlor in environmental waters
Abstract:
S-metolachlor (S-MET) is one of the most widely produced and applied herbicides in China. Owing to its chemical properties, it tends to persist in soil and easily contaminates surface and groundwater through leaching and runoff. This environmental persistence poses a serious threat to plant development and, through the food chain, to human health.
To address the limitations of current detection technologies and meet the growing demand for high-efficiency analytical tools, the researchers employed a mammalian expression system to generate recombinant antibodies targeting S-MET.
Building on the successful expression of these antibodies, they established a sensitive immunoassay for monitoring S-MET residues in various environmental water samples. The icELISA results showed that the recombinant antibodies retained the sensitivity, specificity, and biological activity of the original monoclonal antibodies, delivering accurate and reproducible detection in river water, agricultural runoff, and tap water. View details>>
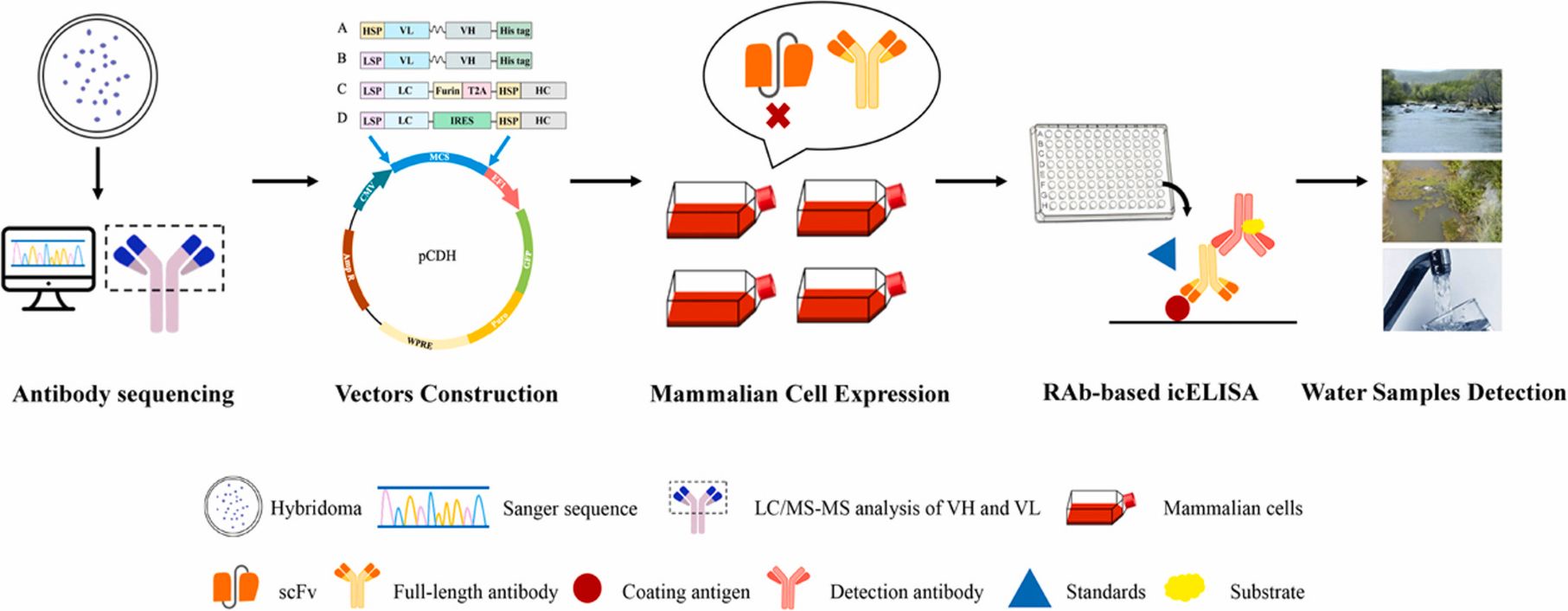
Graphical abstract
Dumbbell probe initiated multi-rolling circle amplification assisted CRISPR/Cas12a for highly sensitive detection of clinical microRNA
Abstract:
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that regulate gene expression by interacting with the mRNAs of target genes. Given their crucial role in the development and progression of various diseases, miRNAs have emerged as promising biomarkers for clinical diagnostics.
In this study, researchers established a novel detection platform, termed DBmRCA, which combines dumbbell probe-initiated multi-rolling circle amplification with the high-sensitivity signal output of CRISPR/Cas12a. This enzyme-free, isothermal method enables accurate quantification of miRNA within just 30 minutes.
Clinical validation revealed that the expression levels of miR-200a and miR-126 were significantly downregulated in lung cancer tissues, and results from DBmRCA were consistent with those obtained by conventional techniques. With its high sensitivity, rapid turnaround, and simplified workflow, the DBmRCA platform presents a reliable tool for miRNA detection and holds strong promise for early diagnosis and therapeutic monitoring of lung cancer. View details>>
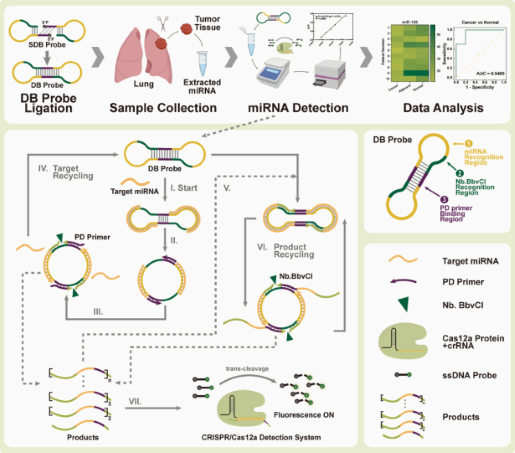
Graphical abstract




 Overexpression Cell Line
Overexpression Cell Line
 Cas9 Expressing Cell Lines
Cas9 Expressing Cell Lines Luciferase Cell Lines
Luciferase Cell Lines CopGFP Cell Lines
CopGFP Cell Lines