 Knockdown Cell Line
Knockdown Cell Line
Service Details
| Cell Types | A wide range of cell types, including tumor cells, standard cell lines, stem cells, primary cells, and immortalized cell lines. |
|---|---|
| Service Types | shRNA-mediated knockdown cell lines / CRISPRi cell lines |
| Deliverables | 1.Gene knockdown polyclonal cell pools (2 vials per line, 1×10⁶ cells per vial) 2.Gene knockdown monoclonal cell lines ≥1 clone (2 vials per clone, 1×10⁶ cells per vial) 3.Project report |
| Cycle/Price |
Delivery in as fast as 6 weeks |
EDI-Service Advantages
Versatile Vector Selection
Efficient Transfection
Precise Clone Screening
Experienced Team
Service Types
shRNA Knockdown Cell Lines
shRNA (short hairpin RNA) stable knockdown cell lines are generated by stably integrating designed shRNA sequences into the host cell genome, creating cell models with long-term and consistent gene silencing. Once expressed in cells, the shRNA is processed by the Dicer enzyme into siRNA (small interfering RNA), which subsequently associates with the RISC (RNA-induced silencing complex). The RISC complex then recognizes and cleaves complementary mRNA molecules, ultimately leading to silencing of the target gene. Contact Us
Service Process
CRISPRi Cell Lines
CRISPR interference (CRISPRi) is a CRISPR-based gene regulation technology that employs a catalytically inactive Cas9 protein (dCas9) combined with a single guide RNA (sgRNA). The dCas9-sgRNA complex binds near the transcription start site of the target gene, creating steric hindrance that represses transcription and inhibits gene expression. In mammalian cells, dCas9 can also be fused with transcriptional repressor domains (e.g., KRAB) for further suppression of gene expression. By designing specific sgRNAs, CRISPRi can simultaneously target multiple genomic loci, enabling coordinated regulation of complex gene networks and providing a powerful tool for functional genomics studies. Contact Us
Service Process
Successful Case
Case 1: Lentivirus-mediated shRNA Knockdown of CHRNA5 Gene Expression
Strategy: A specific short hairpin RNA (shRNA) sequence was designed and synthesized targeting the CHRNA5 gene. The shRNA sequence was cloned downstream of the U6 promoter in the lentiviral vector EDV3021-pLV3-U6-MCS-CopGFP-Puro to construct the shRNA expression plasmid. High-titer lentiviral particles were packaged and used to infect the target cell line. Stable cell lines with integrated shRNA expression cassettes were obtained through puromycin selection. qRT-PCR was performed to assess CHRNA5 mRNA expression levels in the infected cells. The results confirmed that the designed shRNA effectively downregulated CHRNA5 expression (results are shown in the figures/tables below).
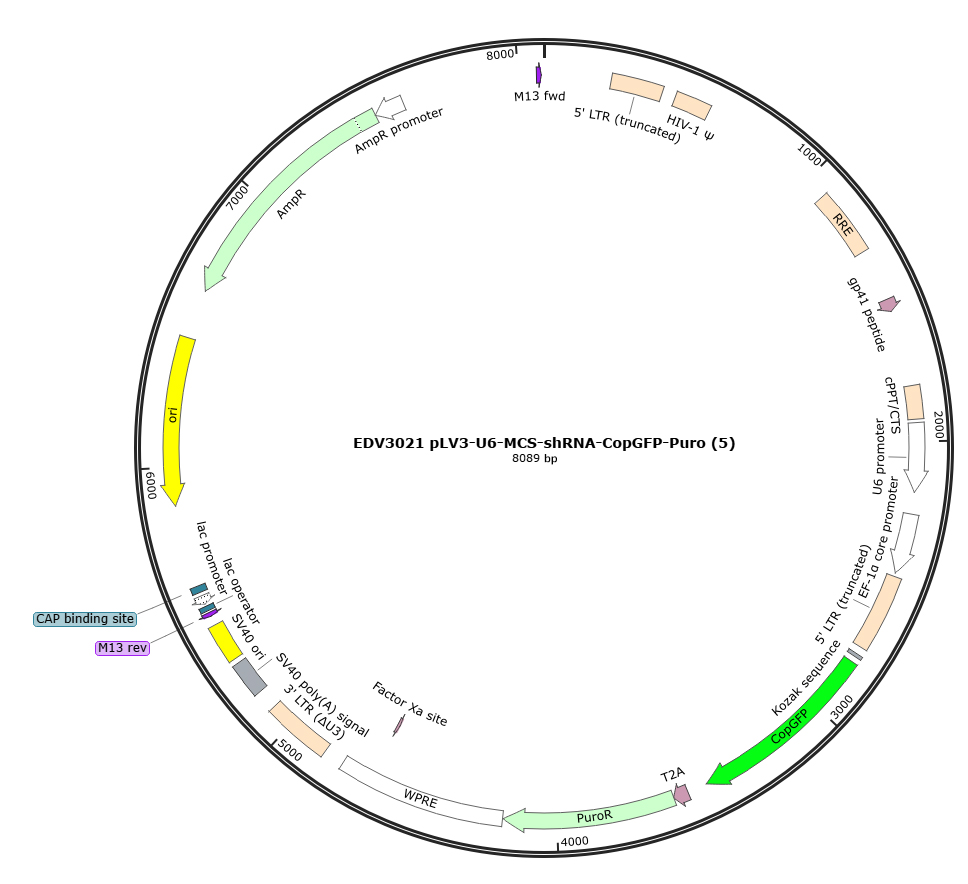
Figure 1: Map of the EDV3021-pLV3-U6-MCS-CopGFP-Puro vector backbone
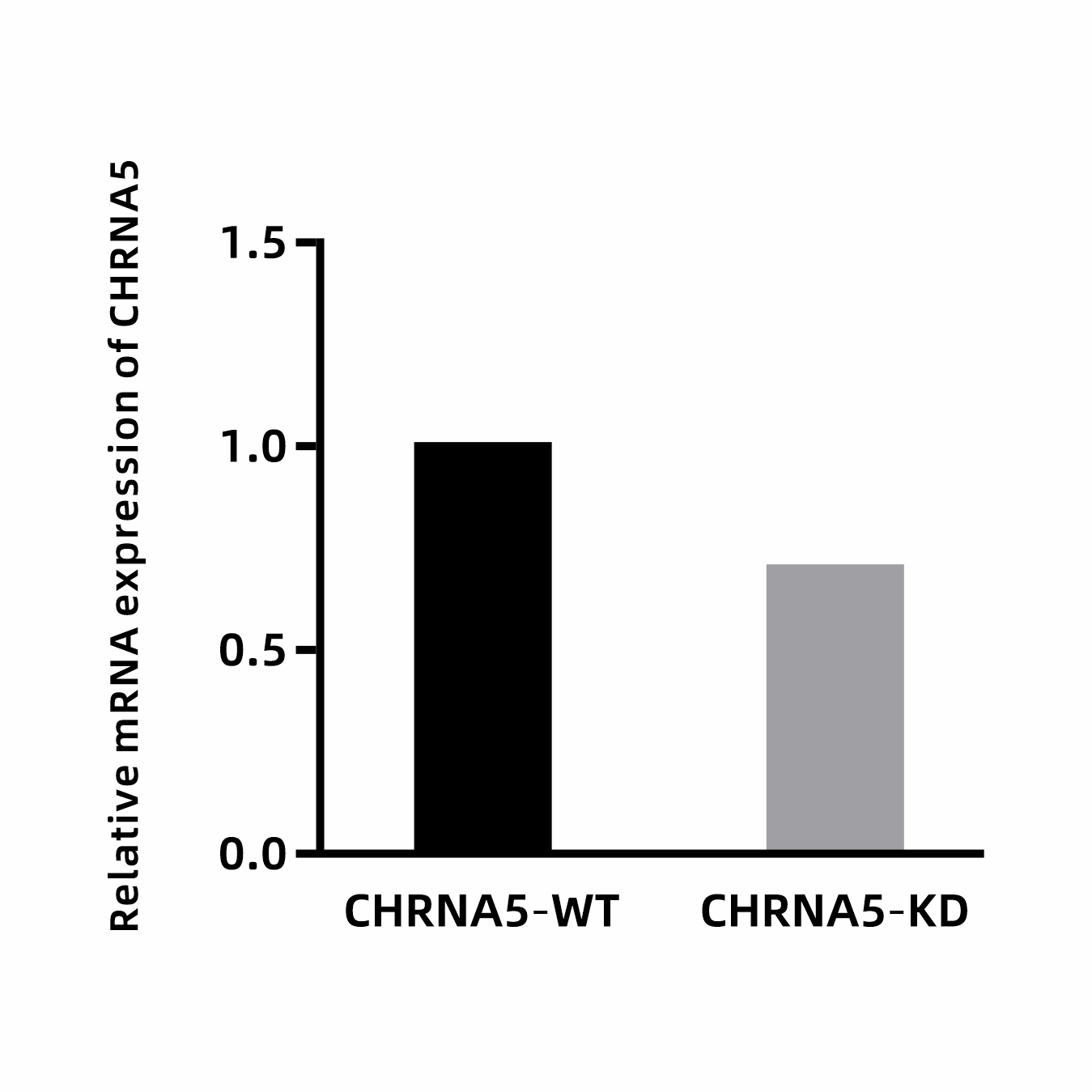
Figure 2: qRT-PCR detection of CHRNA5 gene knockdown
Case 2: Transcriptional Repression of RPL4 Gene Using the CRISPRi System
A lentivirus-mediated CRISPR interference (CRISPRi) system was employed to achieve specific transcriptional repression of the RPL4 gene.
①1.Effector Delivery: The EDITGENE vector EDV2936-pLX_311-KRAB-dCas9 was packaged into high-titer lentiviral particles and used to infect the target cell line. Stable cell lines expressing the KRAB-dCas9 fusion protein were obtained through Blasticidin selection.
②2.Target sgRNA Delivery: Two single guide RNAs (sgRNAs) targeting the promoter/exon regions of RPL4 were designed and cloned into the lentiviral sgRNA expression vector EDV00003-LentiGuide-puro, generating the targeting constructs. Lentiviral particles were packaged and used to infect the above effector-expressing stable cell lines. Stable integration of the sgRNAs was achieved through puromycin selection, resulting in two sgRNA-expressing cell lines.
③3.Detection: qRT-PCR was performed to confirm downregulation of RPL4 at both mRNA and protein levels (results shown in the figures below).
*Note: KRAB-dCas9 recruits chromatin repression complexes to achieve efficient epigenetic silencing without DNA cleavage, enabling potent transcriptional repression of target genes.
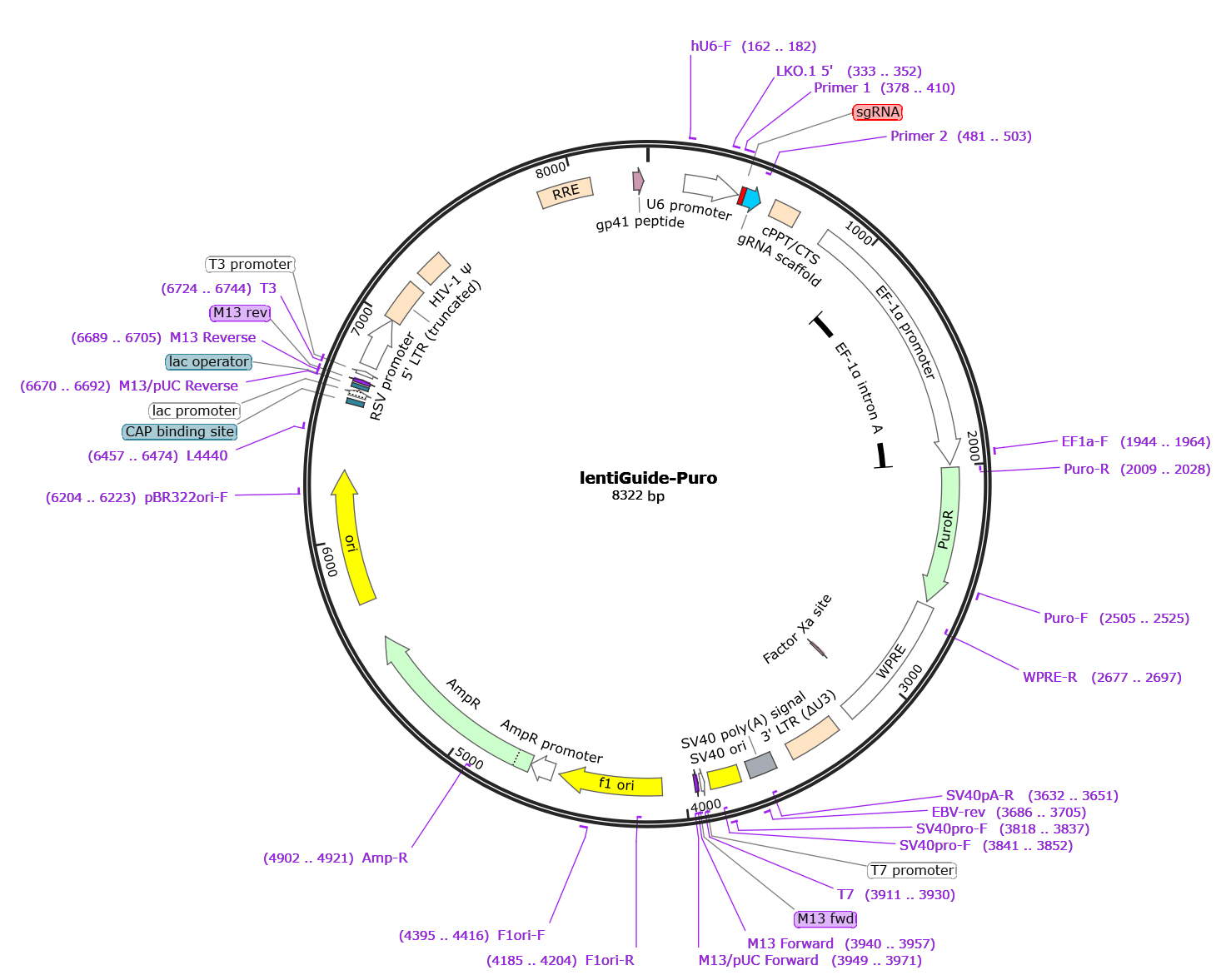
Figure 1: Map of the EDV00003-LentiGuide-puro vector backbone
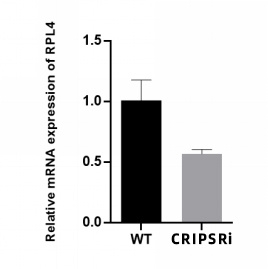
Figure 2: qRT-PCR detection of RPL4 gene knockdown
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
FAQ
Duration of Gene Silencing (Transient Transfection vs. Stable Cell Lines):
| siRNA: | Transient effect (5–7 days),suitable for short-term phenotypic analysis |
|---|---|
| shRNA/CRISPRi: | Stable knockdown after selection, permanent suppression, effective over 20+ passages |
What is Gene Interference and How Does It Work?
What types of gene interference technology services do we mainly provide? (including siRNA transfection, shRNA-mediated lentiviral/adenoviral stable knockdown, and CRISPRi-based transcriptional repression.)
| Technology | Applications | Delivery Format |
|---|---|---|
| siRNA transfection | Rapid validation (effects observed within 72-96 hours) | Cells post-interference/experimental report |
| shRNA viral vectors | Long-term suppression (stable cell line screening) | Viral particles/stable knockdown cell lines |
| CRISPRi | High-specificity gene inhibition | dCas9-expressing cell line and gRNA vector |
Differences Between siRNA, shRNA, and CRISPR Interference (CRISPRi) and How to Choose:
| Parameter | siRNA | shRNA (viral vector) | CRISPRi |
|---|---|---|---|
| Duration of Effect | Transient (5-7 days) | Long-term (stably maintained through passages) | Long-term and stable |
| Cell Type Compatibility | Easily transfected cells | Broad spectrum, including hard-to-transfect cells | Requires pre-established dCas9 cell line |
| Off-target Risk | Medium to high | Medium | Very low |
| Typical Use | Rapid validation | Animal or cell models | High-precision gene regulation |



