 Point Mutation Cell Line
Point Mutation Cell Line
Point mutation cell lines are generated by introducing specific SNP, single-base substitutions or small nucleotide insertions/deletions at specific sites of a target gene. This approach enables precise modeling of disease-associated mutations and facilitates the study of gene function and protein structure. Compared with conventional gene editing methods, point mutations allow refined modifications without disrupting the overall gene architecture, providing researchers with cell models that more closely reflect physiological conditions.
Leveraging our optimized CRISPR/Cas9 and Prime Editing platforms, together with efficient sgRNA design and stringent single-clone screening, EDITGENE offers customized point mutation cell lines of various types. Our services feature editing efficiencies of up to 98%, off-target rates as low as 0.1%, and comprehensive validation, helping accelerate the progress of research projects.Find 100+ Ready-to-Use mutant Cell Lines→
Service Details
| Cell Types | Various cell types, including tumor, conventional, stem, primary, and immortalized cell lines. |
|---|---|
| Services | Prime Editing / Base Editing / HDR |
| Deliverables | Gene point mutation monoclonal cell line ≥ 1 clone (2 vials per clone, 1×10^6 cells per vial) |
| Turnaround/Price |
As fast as 8 weeks,as low as $4500 |
EDI-Service Advantages
Efficient Editing System
Optimized pegRNA Design
Enhanced Cas9n-RT Enzyme
Advanced Transfection System
Streamlined Monoclonal Screening
Experienced Team
Comparison of Point Mutation Methods
| Feature | Prime Editing | Base Editing | HDR |
|---|---|---|---|
| Requirement for DSB | No (single-strand nick only) | No | Yes |
| Editing type | Point mutations, small insertions or deletions | Limited to C→T or A→G conversions | Any type (requires donor template) |
| Efficiency | High | High | Low (<10%; depends on cell division, can be improved with NHEJ inhibitors such as SCR7) |
| Applicable cells | Dividing and non-dividing cells | Dividing and non-dividing cells | Mainly dividing cells |
| Off-target risk | Low | Medium (bystander editing possible) | High (NHEJ-mediated indels) |
Service Types
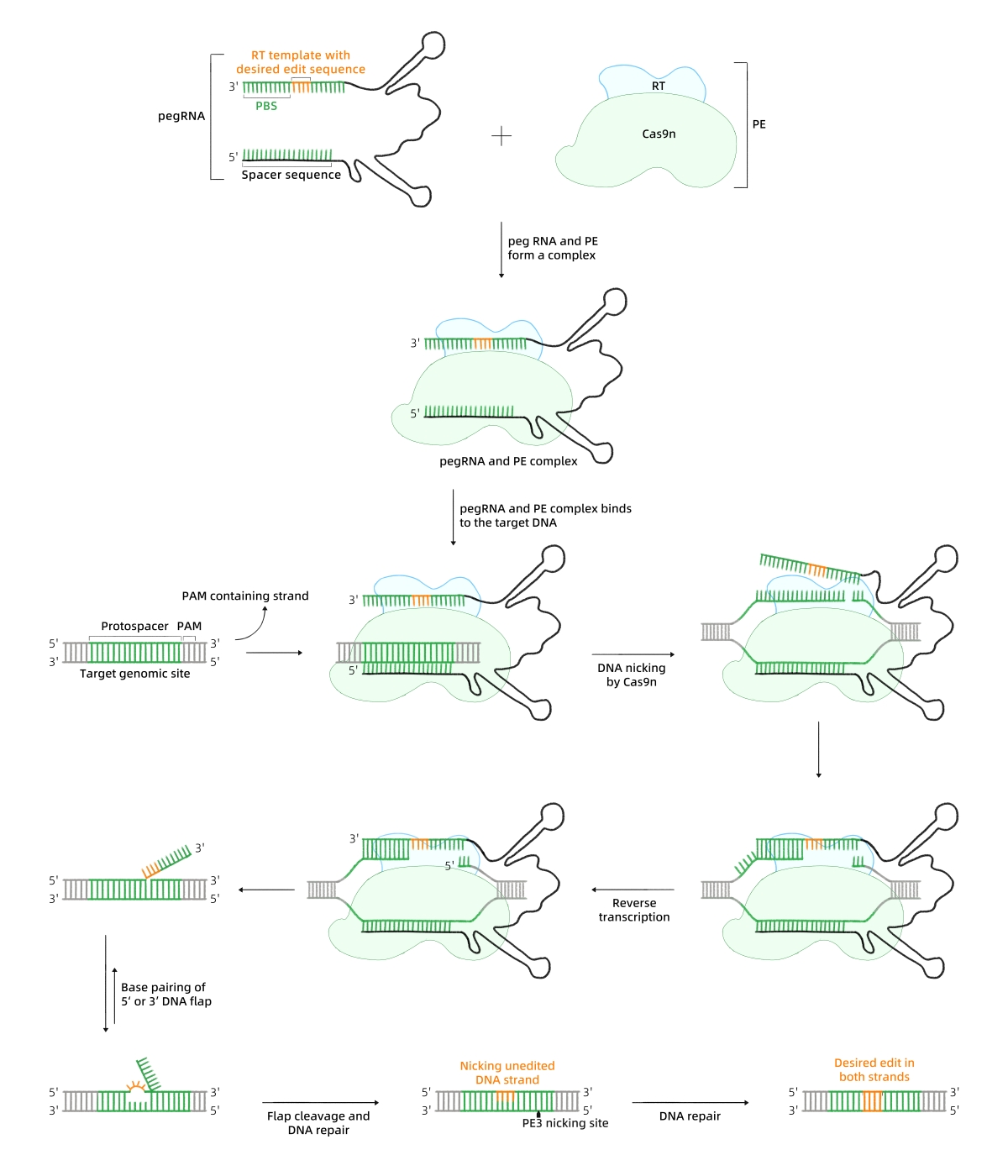
Prime Editing
Working Principle
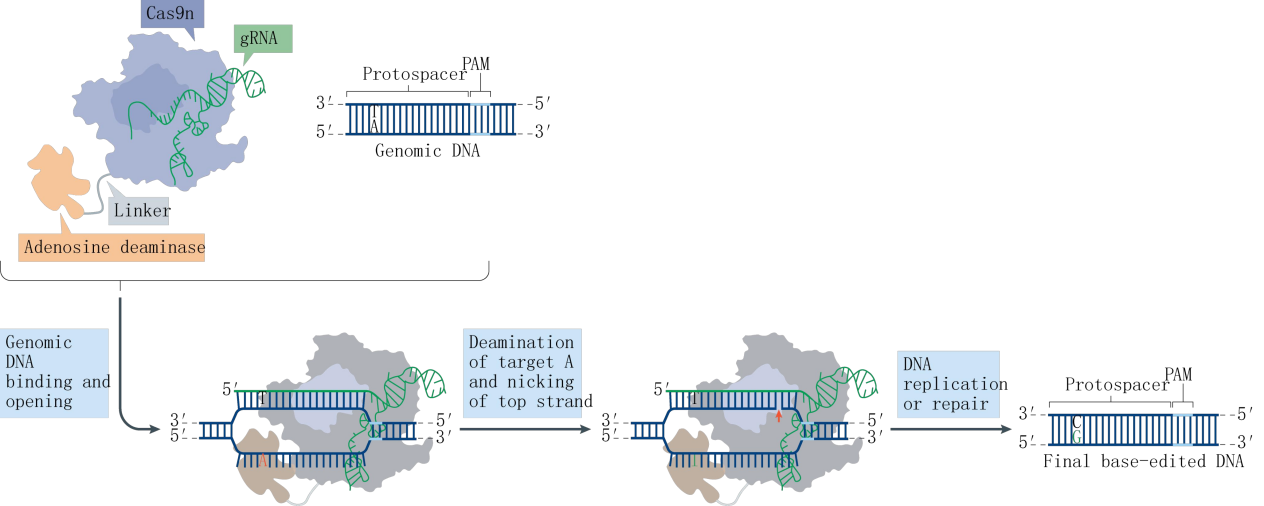
Base Editing
Working Principle
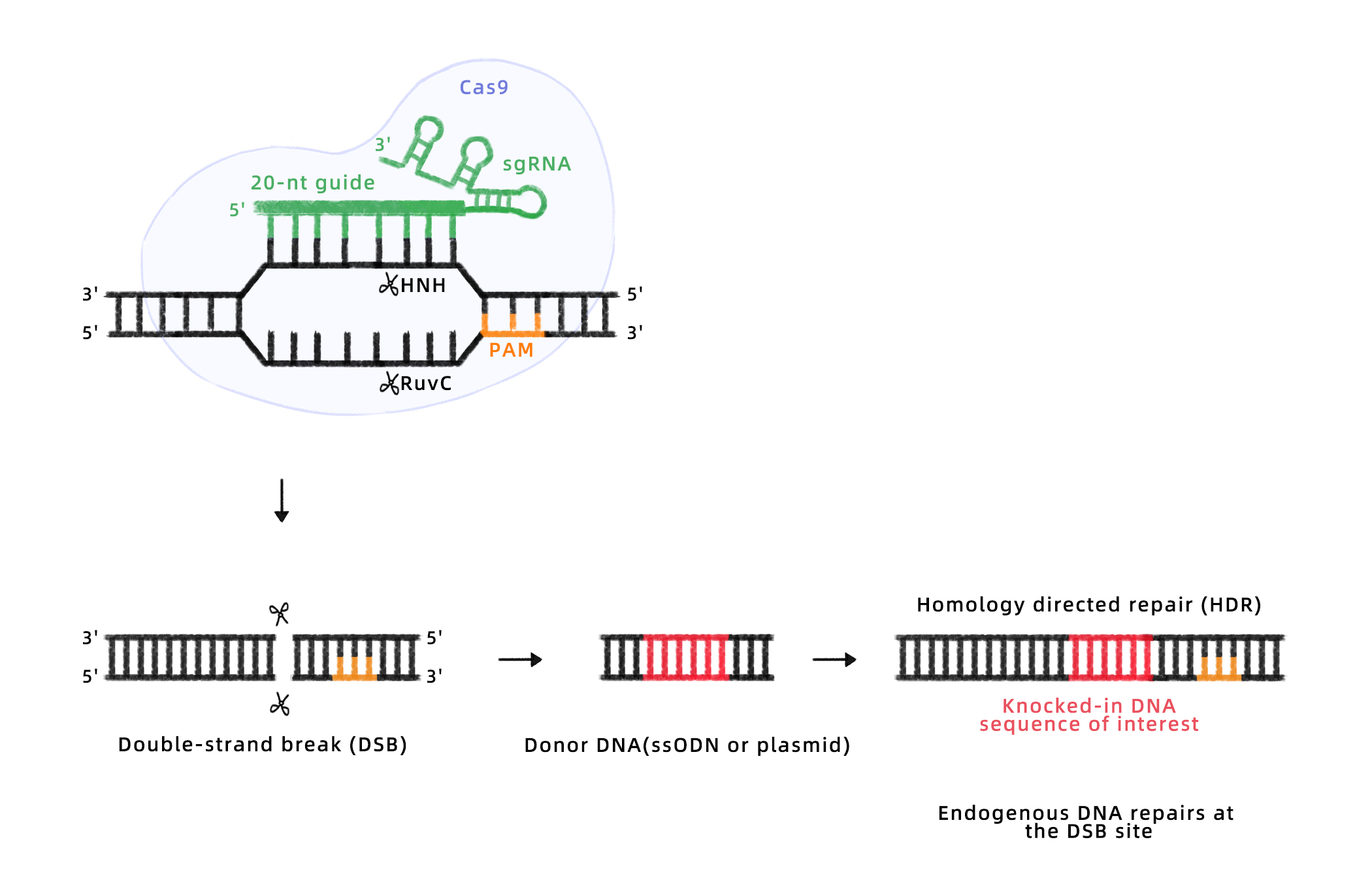
HDR
Working Principle
Case Study
Generation of Single-Base Mutation Cell Lines in A549, K562, and iPSC Cells Using Prime Editing
| Cell type | cell pool editing efficiency | Sequencing peak profile | Number of single clones selected | Number of homozygous single clones |
|---|---|---|---|---|
| A549 | 98% | WT:GAGTTGCGCATTAACAGTGGTGGGA MT:GAGTTGCGCATTAACGGTGGTGGGA 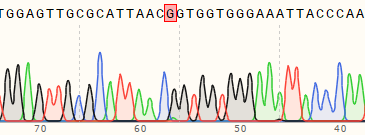 |
7 | 6 |
| K562 | 82% | WT:GTGGAGAAGCCCTTCGGGAGGGACC MT:GTGGAGAAGCCCCTCGGGAGGGACC 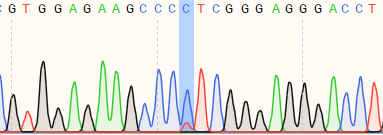 |
6 | 4 |
| IPSC | 60% | WT:AGGGAACCCCAAGTTGAACTTGGCTT MT:AGGGAACCCCAAGTTCAACTTGGCTT 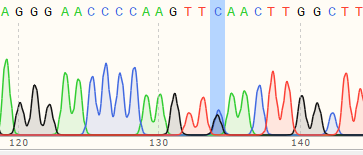 |
8 | 2 |
Generation of Double-Point Mutation Cell Lines in A549 Cells Using Prime Editing
| Cell type | cell pool editing efficiency | Sequencing peak profile | Number of single clones selected | Number of homozygous single clones |
|---|---|---|---|---|
| A549 | 56% and 63% | WT:CAGTCCTCAAAATCTCCATCCCTGT MT:CAGTCCTCAAAACCCCCATCCCTGT 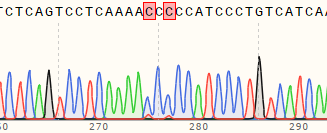 |
4 | 2 |
Generation of Base Insertion Cell Lines in Hela Cells Using Prime Editing
| Cell type | cell pool editing efficiency | Sequencing peak profile | Number of single clones selected | Number of homozygous single clones |
|---|---|---|---|---|
| Hela | 64% | WT:TCCGCTACCACCAATGCCTAATGCATTTGG MT:TCCGCTACCACCAATGCTAATAACTAATGCATTTGG 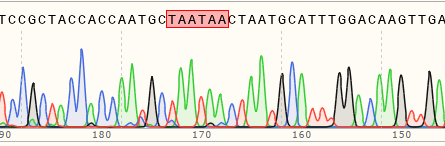 |
11 | 1 |
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Reference Materials
Point mutation of multiple Type 2 Diabetes (T2D) risk SNVs in iPS cells
With advancements in human molecular genetics and genomics, thousands of gene loci associated with common disease risks have been identified, often containing multiple candidate variants. However, most disease-associated variants are non-coding, making it challenging to elucidate the molecular mechanisms underlying these variants. The development of CRISPR technology provides a more precise approach for targeted genome editing, particularly prime editing, which can mediate nearly any single-nucleotide substitution. Consequently, many researchers hope to apply prime editing to study the functional relevance of specific gene variants in pluripotent stem cells, enabling the generation of isogenic cell lines to control genetic background while assessing the dose effect of causal alleles.
In this article, researchers developed an efficient CRISPR prime editing technology to generate cell lines carrying heterozygous or homozygous alleles in induced pluripotent stem cells (iPSCs) and optimized the prime editing technique. Six single-nucleotide variants (SNVs) associated with Type 2 Diabetes (T2D) risk were selected for editing, and iPSCs derived from human donors with different genetic backgrounds were edited at each site. The researchers successfully generated 27 edited iPSC clones covering 6 SNVs associated with T2D or congenital hyperinsulinemia (CHI) and found that prime editing was more efficient in iPSCs than in HEK293T cells. Overall, this study demonstrates the potential of prime editing for generating iPSCs with specific genetic backgrounds, providing a powerful tool to study the effects of specific genetic variants on disease.
Point mutation of the HBBS gene in hematopoietic stem and progenitor cells (HSPCs)
Sickle-cell disease (SCD) is an autosomal recessive genetic disorder caused by an A·T to T·A point mutation in the β-globin gene (HBB). Currently, the only FDA-approved cure for SCD is allogeneic hematopoietic stem cell transplantation; however, most patients lack ideal donors, and this procedure can lead to severe toxicity. Correcting a patient’s own hematopoietic stem cells (HSCs) can bypass immune complications and eliminate the need for matched donors. Clinical trials are ongoing to correct the SCD mutation using Cas9 nuclease-initiated homology-directed repair (HDR) and adeno-associated virus type 6 (AAV6)-delivered DNA templates.
In this article, researchers applied an optimized prime editing system to correct HSPCs from SCD patients ex vivo. By electroporating PEmax mRNA along with synthetic epegRNA and nicking sgRNA, the SCD allele (HBBS) was successfully corrected back to the wild-type (HBBA), with correction rates between 15% and 41%. Subsequently, edited HSPCs were transplanted into immunodeficient mice, and 7 weeks later, edited HSPCs maintained HBBA levels in the bone marrow, with engraftment rates, hematopoietic differentiation, and lineage maturation similar to unedited healthy donor HSPCs. Therapeutic evaluation revealed that, post-transplant, 42% of red blood cell precursors and reticulocytes expressed HBBA, exceeding the predicted therapeutic benefit level. Gene-specific analyses also confirmed high DNA specificity of the prime editing system. The study demonstrates the potential of prime editing in SCD treatment, showing its effectiveness in improving therapeutic outcomes while reducing off-target editing risks.
Iterative upgrades to prime editing technology by studying Prime editor’s mechanism of action
This article elucidates the structural basis of pegRNA-guided reverse transcription in prime editing technology. High-resolution structural analysis revealed the three-dimensional conformation of key proteins in the Prime Editor, including Cas9 and reverse transcriptase, and their interaction with pegRNA. It details how pegRNA guides the Prime Editor to recognize specific DNA sequences and perform reverse transcription, inserting a predetermined gene sequence into the target site. The study also discusses the roles of key amino acid residues in pegRNA binding and reverse transcription, providing insight into the precise mechanisms of these molecular interactions. This finding not only deepens our understanding of the Prime Editing mechanism but also offers valuable structural information for optimizing and improving this technology, advancing its application in gene therapy and other fields.
Increasing Prime editing efficiency via truncated RT enzyme with AAV delivery
Prime editing is a novel CRISPR-based genome-editing technology that does not require double-strand DNA breaks or exogenous donor template DNA, showing great potential in biomedical research and gene therapy. Despite its versatility and precision, prime editing efficiency varies across different editing types, target sites, and cell types. Therefore, to expand its applications, there is a need to improve prime editing efficiency.
Researchers screened 11 different RT variants, optimized with GenScript algorithm for human codons, which increased PE protein expression levels by 1.4-fold. By deleting the RNase H domain and further shortening the RT sequence, they created multiple truncated PECO variants, reducing the Prime editor length by 621 bp without compromising editing efficiency. To enable efficient dual-AAV delivery of PE, the team constructed a split PE system based on different Cas9 cleavage sites and inteins, identifying cleavage sites Rma 573-574 and 674-675. When these sites were paired with Rma intein, they significantly increased AAV vector titers and Prime editor efficiency. Through engineering and optimization, researchers successfully enhanced Prime editing efficiency and resolved AAV size constraints, providing a more efficient tool for future gene therapy and biomedical research.
Cryo-EM study of Prime editor complex to facilitate iterative upgrades to prime editing
The molecular mechanism of how the Prime editor recognizes pegRNA and interacts with target DNA remains unclear, limiting the understanding and optimization of the prime editing process. To address this, researchers determined the cryo-electron microscopy (cryo-EM) structure of the Prime editor in various states, providing a structural framework for understanding this innovative genome engineering system.
Using cryo-EM technology, researchers analyzed the Prime editor complex’s structure in different states, including pre-initiation, initiation, extension, and termination, successfully obtaining high-resolution structures that reveal dynamic changes in the reverse transcription guidance process. Based on structural information, researchers designed pegRNA and Prime editor variants, truncating and fusing M-MLV RT to create a smaller Prime editor variant (PECO-Mini) that maintained editing efficiency while increasing AAV vector titers and prime editing efficiency. Activity testing results showed that the engineered Prime editor variant has comparable activity to the original Prime editor in vitro. This study reveals the structural characteristics of the Prime editor complex in various working states, providing key information for understanding its molecular mechanism.
Selected Customer Resources
Deep whole-genome analysis of 494 hepatocellular carcinomas
Abstract:
To date, more than half of global hepatocellular carcinoma (HCC) cases occur in China, yet comprehensive whole-genome analyses focusing on HBV-related HCC within the Chinese population remain scarce. To address this challenge, researchers initiated the China Liver Cancer Atlas (CLCA) project, aiming to conduct large-scale whole-genome sequencing to unravel the unique pathogenic mechanisms and evolutionary trajectories of HCC in China.
The researchers performed deep whole-genome sequencing on 494 HCC tumor samples, with an average depth of 120×, alongside matched blood controls, providing a detailed genomic landscape of HBV-associated HCC. Beyond confirming well-known coding driver genes such as TP53 and CTNNB1, the study identified six novel coding drivers—including FGA—and 31 non-coding driver genes.
Additionally, the research uncovered five new mutational signatures, including SBS_H8, and characterized the presence of extrachromosomal circular DNA (ecDNA) formed via HBV integration, which contributes to oncogene amplification and overexpression. Functional validation experiments demonstrated that mutations in genes such as FGA, PPP1R12B, and KCNJ12 significantly enhance HCC cell proliferation, migration, and invasion.
These findings not only deepen our insights into the genomics of HCC, but also open up new potential targets for diagnosis and therapy. View details>>
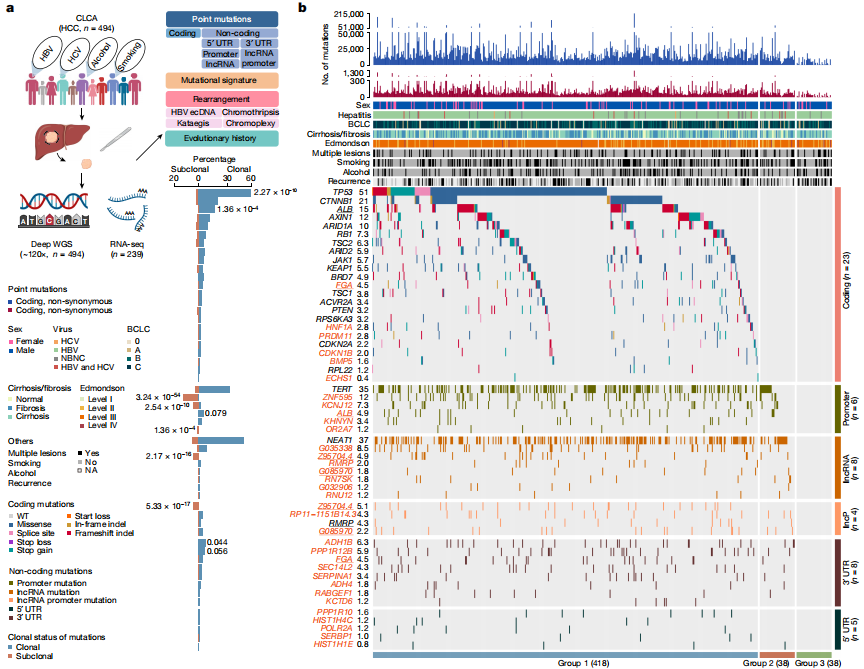
Candidate driver landscape
Targeted Macrophage CRISPR-Cas13 mRNA Editing in Immunotherapy for Tendon Injury
Abstract:
During the acute inflammatory phase of tendon injury, excessive activation of macrophages leads to the overexpression of SPP1, which encodes osteopontin (OPN), thereby impairing tissue regeneration. The CRISPR-Cas13 system holds great promise for tissue repair due to its unique RNA editing and rapid degradation capabilities; however, its application has been limited by the lack of efficient delivery methods.
To address this, the researchers systematically screened various cationic polymers targeting macrophages and developed a nanocluster carrier capable of efficiently delivering Cas13 ribonucleoprotein complexes (Cas13 RNPs) into macrophages. Utilizing a reactive oxygen species (ROS)-responsive release mechanism, this system specifically suppresses the overexpression of SPP1 in macrophages within the acute inflammatory microenvironment of tendon injury.
Experimental results demonstrated that this targeted delivery strategy significantly reduced the population of SPP1-overexpressing macrophages induced by injury, inhibited fibroblast activation, and alleviated peritendinous adhesion formation. Furthermore, the study elucidated that SPP1 promotes fibroblast activation and migration through the CD44/AKT signaling pathway, and that inhibiting this pathway effectively mitigates adhesion formation following tendon injury. View details>>
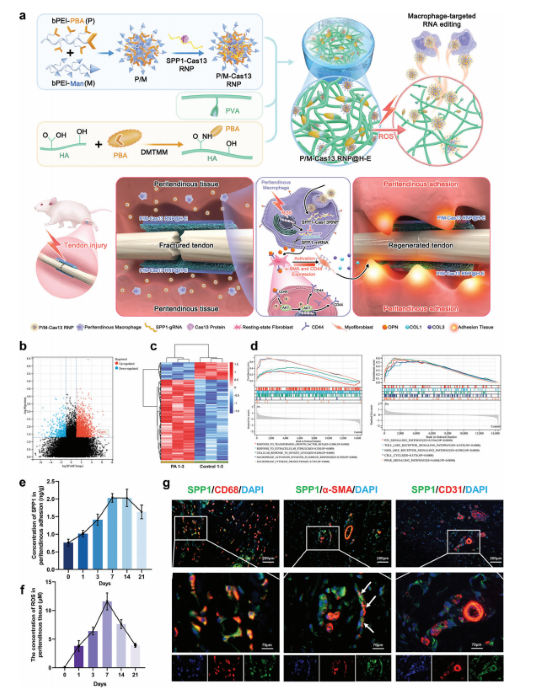
Schematic diagram illustrating immune microenvironment-activated mRNA editing strategies of macrophages for PA therapy
Electrical stimulation of piezoelectric BaTiO3 coated Ti6Al4V scaffolds promotes anti-inflammatory polarization of macrophage and bone repair via MAPK/JNK inhibition and OXPHOS activation
Abstract:
Spinal cord injury (SCI) is a severe disabling condition that causes permanent loss of sensory, autonomic, and motor functions. While stem cell therapies, particularly mesenchymal stem cells (MSCs), show great promise for SCI treatment, their limited regenerative capacity restricts their application in tissue repair. The researchers observed that extracellular vesicles derived from antler bud progenitor cells (EVsABPC) may carry bioactive signals that promote tissue regeneration. Accordingly, they isolated and engineered EVs from ABPCs for SCI therapeutic investigation.
The study found that EVsABPC significantly enhanced neural stem cell (NSC) proliferation, promoted axonal growth, reduced neuronal apoptosis, and modulated inflammation by shifting macrophage polarization from the pro-inflammatory M1 phenotype to the anti-inflammatory M2 phenotype. Moreover, engineered EVsABPC modified with cell-penetrating peptides demonstrated improved targeting to the SCI lesion site, markedly enhancing neural regeneration and functional motor recovery. These findings highlight EVsABPC as a promising candidate for SCI therapy. View details>>
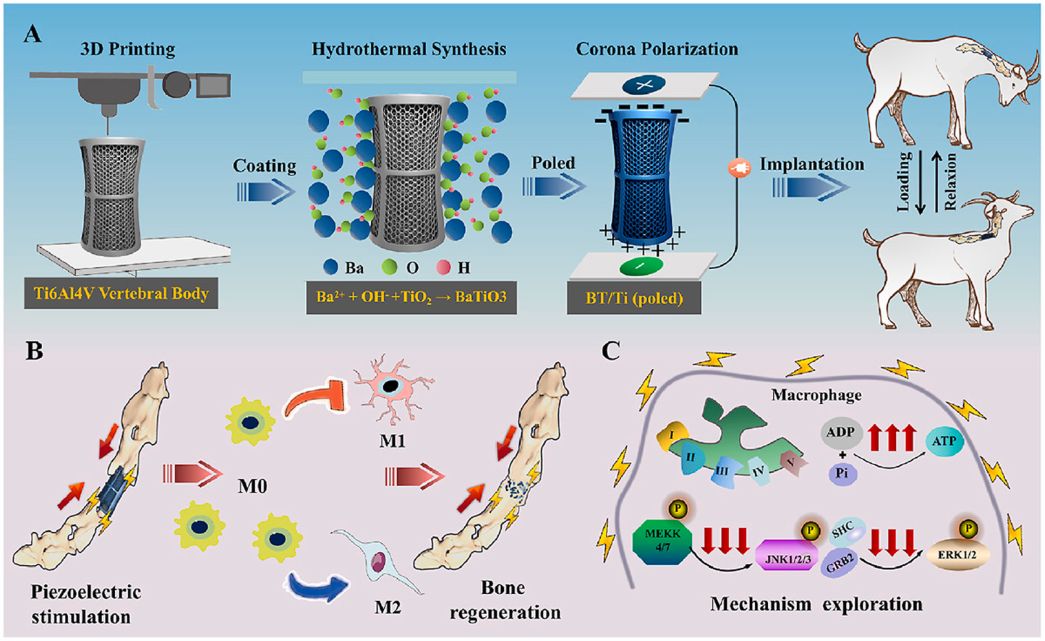
Graphical abstract
Generation of recombinant antibodies by mammalian expression system for detecting S-metolachlor in environmental waters
Abstract:
S-metolachlor (S-MET) is one of the most widely produced and applied herbicides in China. Owing to its chemical properties, it tends to persist in soil and easily contaminates surface and groundwater through leaching and runoff. This environmental persistence poses a serious threat to plant development and, through the food chain, to human health.
To address the limitations of current detection technologies and meet the growing demand for high-efficiency analytical tools, the researchers employed a mammalian expression system to generate recombinant antibodies targeting S-MET.
Building on the successful expression of these antibodies, they established a sensitive immunoassay for monitoring S-MET residues in various environmental water samples. The icELISA results showed that the recombinant antibodies retained the sensitivity, specificity, and biological activity of the original monoclonal antibodies, delivering accurate and reproducible detection in river water, agricultural runoff, and tap water. View details>>
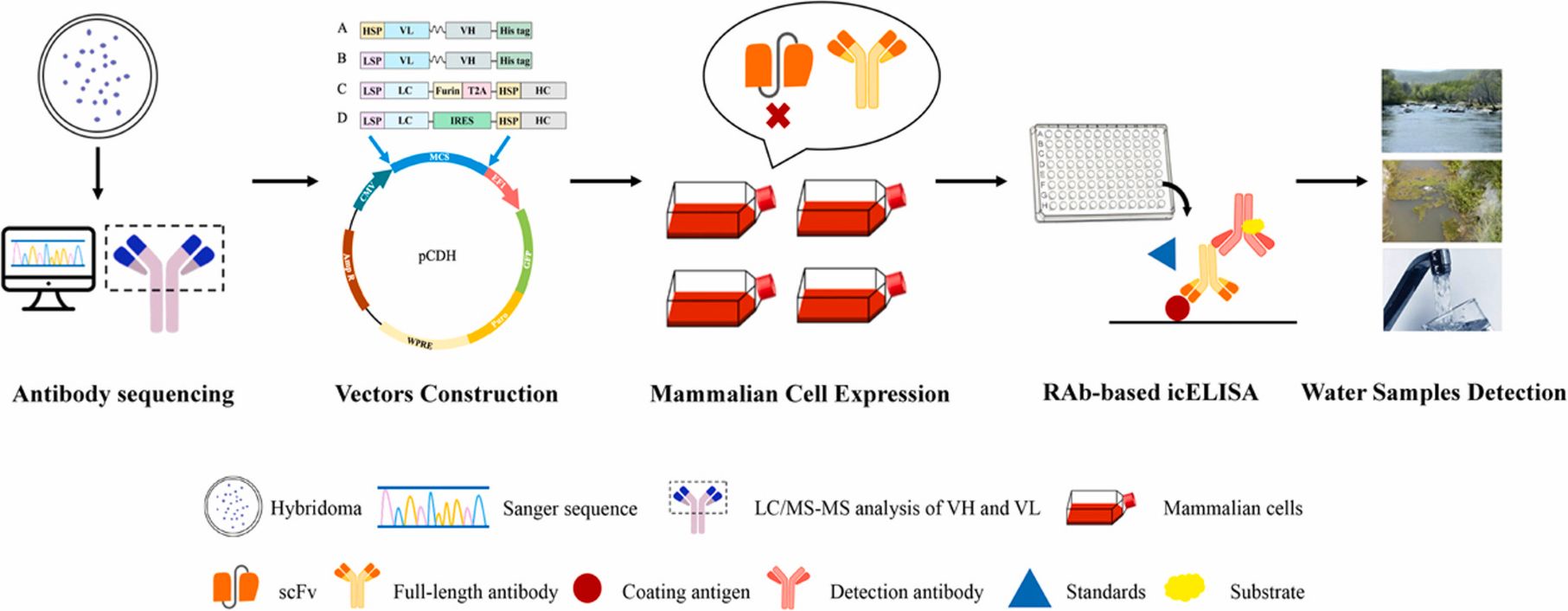
Graphical abstract
Dumbbell probe initiated multi-rolling circle amplification assisted CRISPR/Cas12a for highly sensitive detection of clinical microRNA
Abstract:
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that regulate gene expression by interacting with the mRNAs of target genes. Given their crucial role in the development and progression of various diseases, miRNAs have emerged as promising biomarkers for clinical diagnostics.
In this study, researchers established a novel detection platform, termed DBmRCA, which combines dumbbell probe-initiated multi-rolling circle amplification with the high-sensitivity signal output of CRISPR/Cas12a. This enzyme-free, isothermal method enables accurate quantification of miRNA within just 30 minutes.
Clinical validation revealed that the expression levels of miR-200a and miR-126 were significantly downregulated in lung cancer tissues, and results from DBmRCA were consistent with those obtained by conventional techniques. With its high sensitivity, rapid turnaround, and simplified workflow, the DBmRCA platform presents a reliable tool for miRNA detection and holds strong promise for early diagnosis and therapeutic monitoring of lung cancer. View details>>
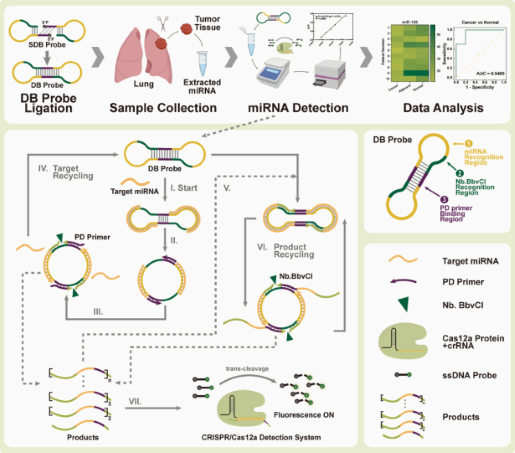
Graphical abstract




