[Literature Review] A Novel NovaIscB System Enables Durable and Efficient Genome Editing
Cas9
In May 2025, Nature Biotechnology published an online research article titled "Evolution-guided protein design of IscB for persistent epigenome editing in vivo."
Led by Chinese scientist Zhang Feng and his team, the study integrated evolution-guided protein engineering strategies to develop NovaIscB, a novel and compact genome editor. The researchers further established a durable epigenetic editing system named OMEGAoff, delivered via a single AAV vector. This breakthrough offers a powerful tool for advancing gene therapies targeting chronic diseases.
Original link: DOI: 10.1038/s41587-025-02655-3
Spotlight
1. 100-Fold Increase in Efficiency:
A highly active IscB variant, OrufIscB, was identified through homologous gene screening. Following engineering of its REC domain, the newly developed NovaIscB demonstrated a 100-fold improvement in editing efficiency over the wild-type, reaching up to 40% indel efficiency.
2. Enhanced Targeting Precision:
The guide RNA length was successfully extended from 13 bp to 20 bp, significantly improving targeting specificity and reducing off-target effects compared to existing compact editors.
3. Single-Vector In Vivo Delivery:
By fusing a methyltransferase domain, the researchers constructed the OMEGAoff system. NovaIscB, together with a truncated ωRNA, can be efficiently packaged into a single AAV vector, overcoming the delivery limitations of large genome editors.
4. Sustained Gene Silencing:
A single in vivo injection of OMEGAoff in mice led to persistent gene silencing for over 6 months, with significantly reduced serum cholesterol levels and no observed hepatotoxicity.
Traditional CRISPR-Cas genome editors are often too large for efficient delivery via adeno-associated virus (AAV) vectors and face challenges in balancing editing efficiency with low off-target effects. Although IscB—an ancestral protein of Cas9 —offers the advantage of a compact size, it suffers from low editing efficiency and requires short guide RNAs. To overcome these limitations, researchers aimed to develop a highly efficient and precise miniature genome editor by leveraging natural evolutionary diversity and protein engineering strategies.
1. Homolog Screening
The researchers screened 144 IscB homologs and 6 type II-D Cas9 homologs, identifying a highly efficient variant named OrufIscB. This variant achieved an editing efficiency of 8%, representing a tenfold improvement over earlier versions.
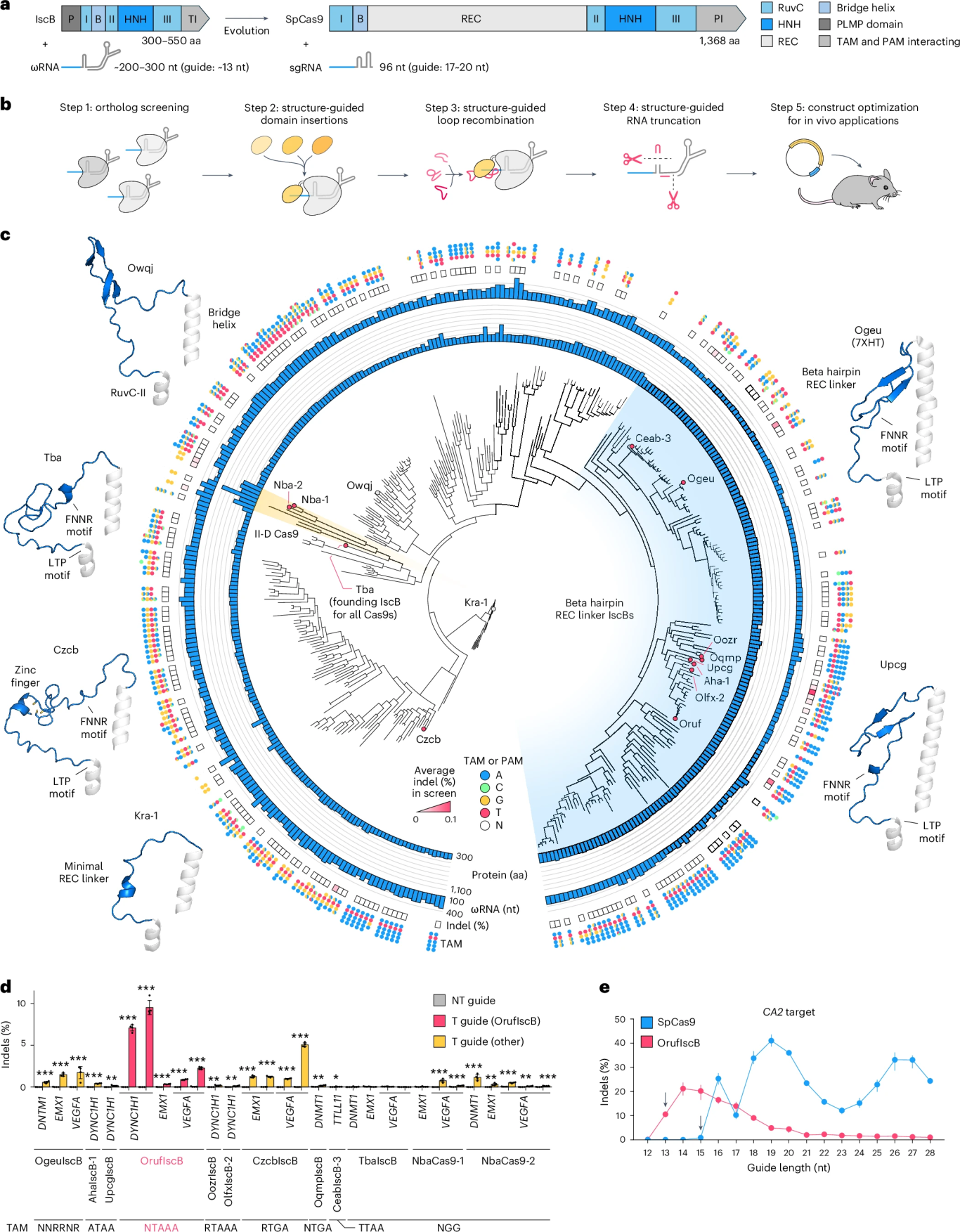
Figure 1. Screening of Natural IscB Variants
2. REC Domain Chimerism
The researchers inserted the REC recognition domain from Cas9 into a conserved region of OrufIscB. Among 183 chimeric constructs, the Nba-1 REC variant was selected for its enhanced performance, extending the effective guide RNA length to 15 bp.
3. Optimization of Key Loop Regions
Through 54 different REC loop-swapping combinations, the researchers ultimately developed NovaIscB, which supports an extended guide RNA length of 20 bp and exhibits a 200-fold improvement in specificity compared to the wild-type IscB.
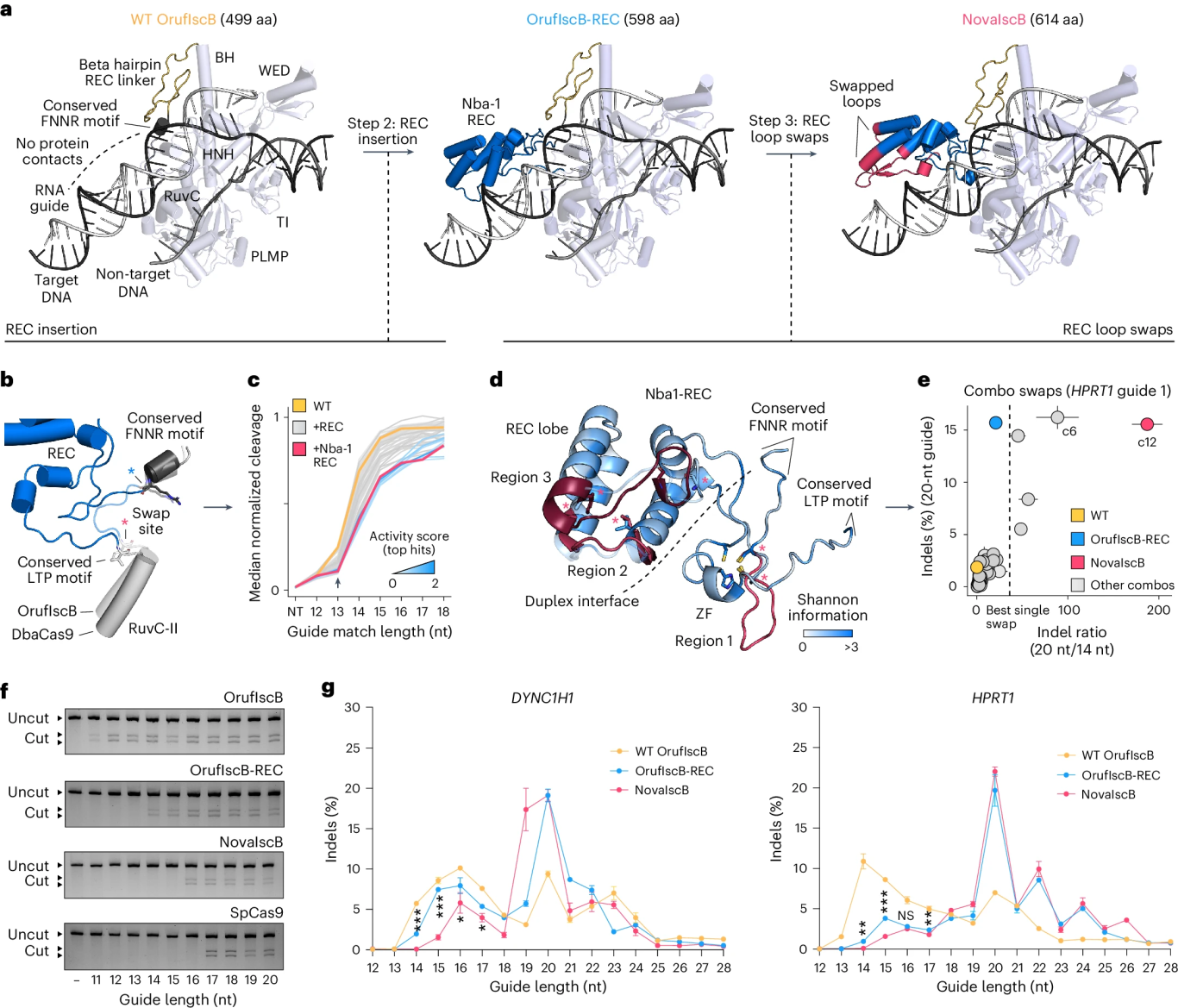
Figure 2. Rational Redesign of OrufIscB for Enhanced Editing Efficiency
The researchers removed non-essential stem-loop structures from the 5' (42 nt) and 3' (17 nt) ends of the ωRNA, reducing its total length from 225 nucleotides to 166 nucleotides. The truncated ωRNA exhibited a fourfold increase in expression levels in human cells.
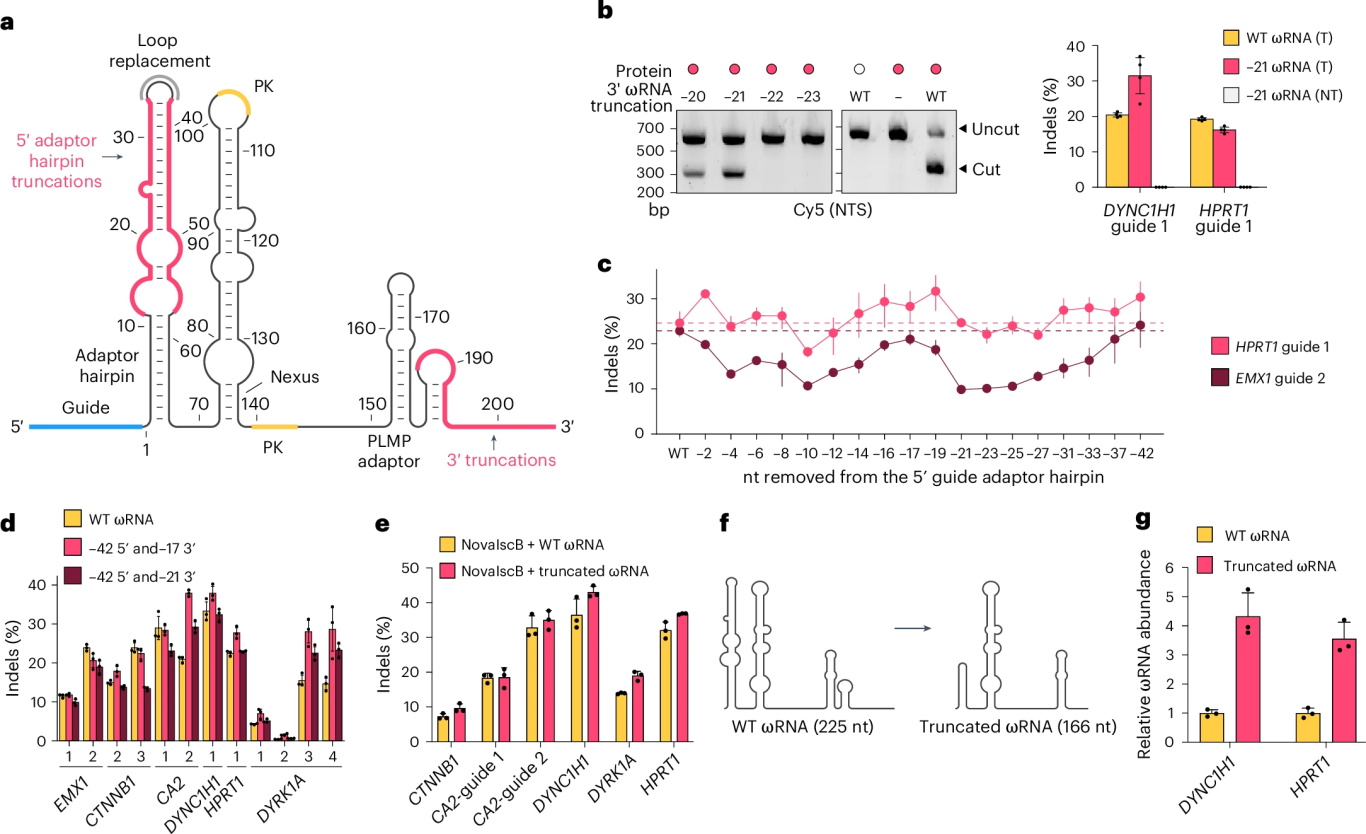
Figure 3. Structure-Guided Engineering of the ωRNA Scaffold
By fusing a catalytically inactive version of OrufIscB-KRK (with mutations in the RuvC and HNH nuclease domains) to DNA methyltransferase Dnmt3A-3L and the transcriptional repressor domain KRAB, the researchers developed a compact epigenetic editing tool named OMEGAoff. In both HEK293FT and AML12 cells, OMEGAoff achieved transcriptional repression comparable to CRISPRoff. Through optimization of the fusion architecture, target gene expression was suppressed to 20–30% of the control level.
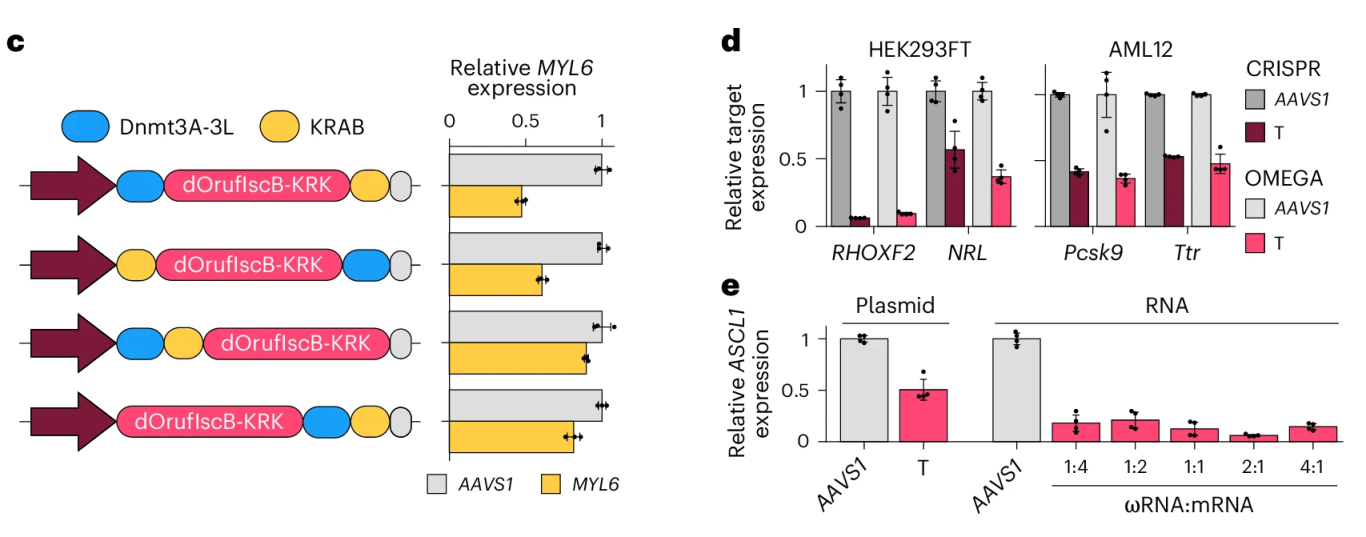
Figure 4. OMEGAoff Achieves CRISPRoff-Comparable Transcriptional Repression
In an in vivo study, mice were intravenously injected with AAVs encoding OMEGAoff and Pcsk9/Rosa26-targeting guides at a dose of 2×10¹¹ total viral genomes per animal. Blood samples were collected at various time points to isolate serum for measuring PCSK9 protein and cholesterol levels.
Results showed that three weeks post-injection, serum PCSK9 levels in the targeted group were reduced by 70%, and cholesterol levels dropped by 40%. These effects persisted for up to six months. Importantly, no elevation of hepatic injury markers (bilirubin/ALT) was observed, indicating good safety and tolerability of the treatment.
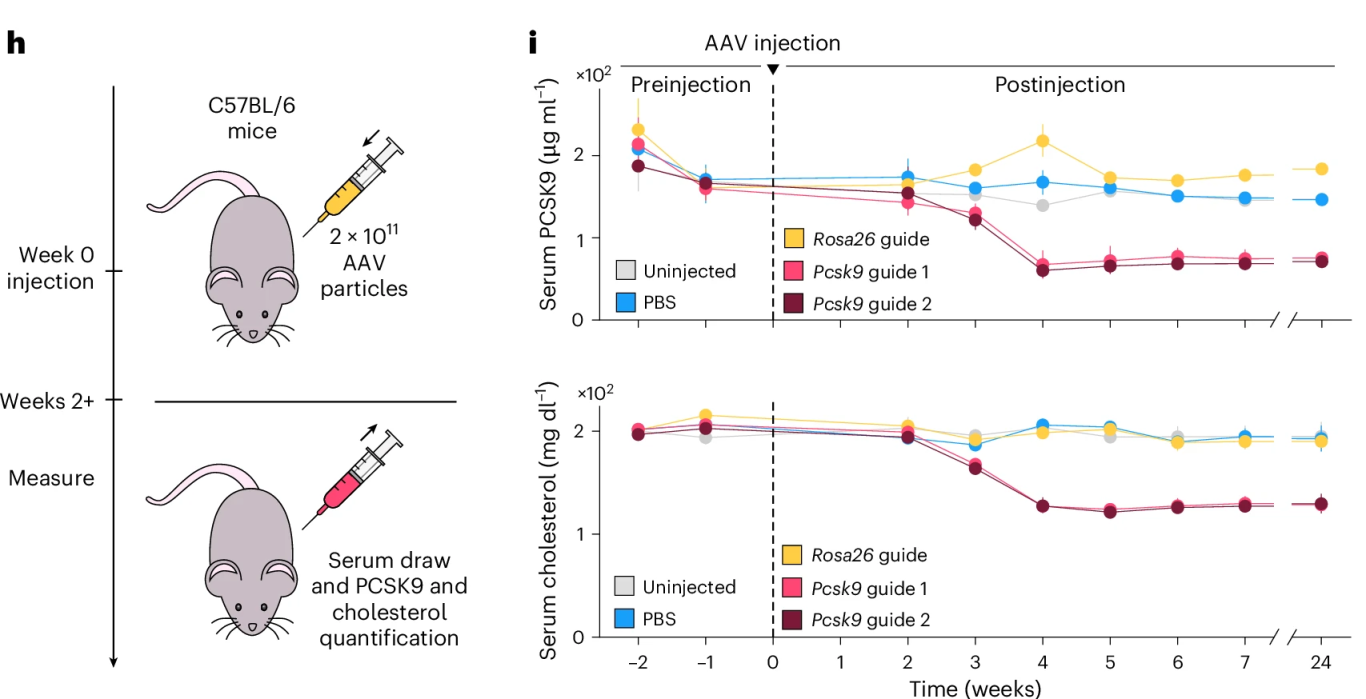
Figure 5. In Vivo Pcsk9 Suppression via AAV-Delivered OMEGAoff Experimental Design and Outcome Analysis
Through an evolution-guided protein engineering strategy, the researchers successfully developed a compact RNA-guided nuclease, NovaIscB. When fused to epigenetic regulators, the resulting editor—OMEGAoff—enabled long-term gene regulation via single-AAV delivery. In vivo experiments demonstrated sustained target gene suppression for no less than 6 months, significant reductions in serum cholesterol levels, and no detectable hepatotoxicity.
These findings highlight the therapeutic potential of NovaIscB and OMEGAoff for the treatment of chronic diseases, offering a groundbreaking platform for compact, safe, and durable gene therapy solutions.















![[Literature Review] The Secret Behind Bone “Hollowing”? STOM Protein's Unexpected Function Uncovered](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)