CRISPR–Csm: Real-time Single-Molecule Tracking of RNA Dynamics in Live Cells
RNP

RNA is crucial for protein synthesis and gene regulation, and understanding its spatiotemporal dynamics is essential for deciphering cellular functions. However, existing imaging techniques face limitations, such as interference from exogenous labeling, detection issues for low-abundance RNA, and inadequate single-molecule tracking precision.
To address these challenges, Chenglong Xia and his team recently introduced an innovative solution, smLiveFISH, published in Nature Biotechnology. This method combines the CRISPR-Csm complex’s precise targeting with a multiple crRNA signal amplification strategy, enabling real-time imaging of unmodified endogenous RNA at the single-molecule level.
Compared to traditional methods, smLiveFISH offers three key advantages:
① Non-invasive, in situ detection that preserves RNA’s natural state.
② Improved visualization of low-abundance RNA.
③ High sensitivity in primary cells like neurons.
This breakthrough technology provides a powerful tool for studying RNA dynamics and regulation in live cells.
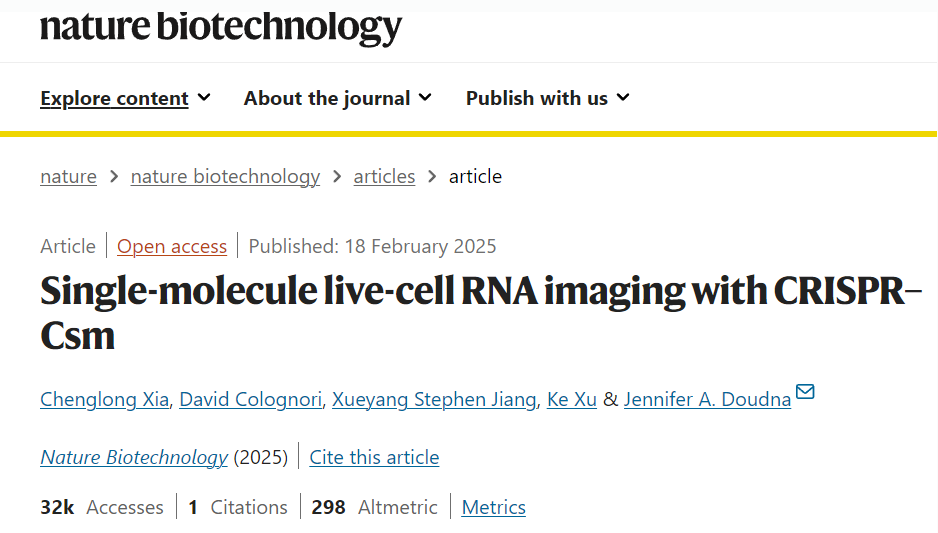
I. CRISPR-Csm Complex Combined with Multiple crRNAs for Precise Targeting of RNA
The research team utilized the Type III-A CRISPR-Csm system as the core tool due to its high specificity in recognizing RNA, enabling single-molecule RNA imaging. In their experiment, they first constructed plasmids encoding the Csm complex and the CRISPR guide array. The Csm complex consists of Csm1, Csm2, dCsm3-2×GFP (a catalytically inactive Csm3 fused with dual GFP), Csm4, Csm5, and Cas6 subunits. Cas6 is responsible for processing the precursor crRNA (pre-crRNA) into mature crRNA, which then assembles into a ribonucleoprotein (RNP) complex with the Csm complex.
To enhance imaging signals, the researchers designed multiple crRNAs, allowing several Csm complexes to "decorate" the target RNA molecule, significantly improving the signal-to-noise ratio and ensuring single-molecule resolution.
For experimental validation, the team selected two mRNAs, NOTCH2 and MAP1B, as target sequences. NOTCH2 mRNA, enriched near the endoplasmic reticulum, was chosen to study co-translation translocation mechanisms, while MAP1B mRNA, associated with axonal growth, was ideal for investigating directional transport due to its enrichment at the cell periphery. The team designed 48 complementary crRNAs for each mRNA and introduced the plasmids into cells through transient transfection. After 48 hours, they successfully observed the dynamic behavior of single RNA molecules using fluorescence microscopy.
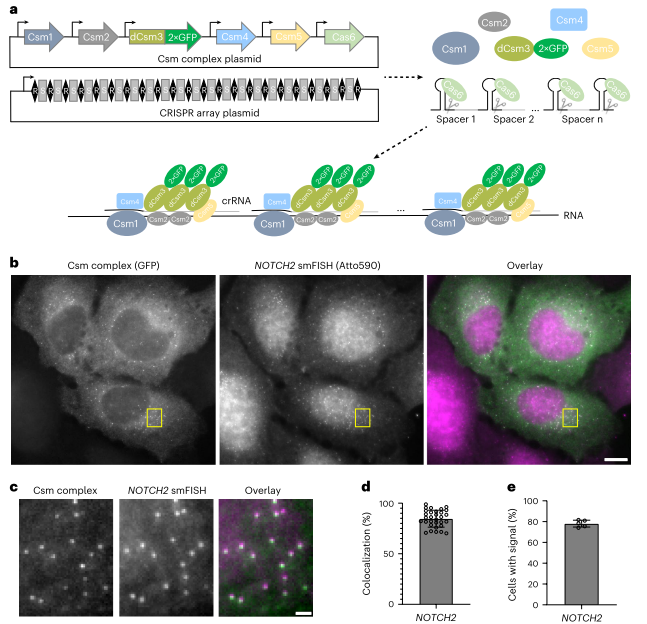
Imaging Natural Single mRNA Molecules with smLiveFISH
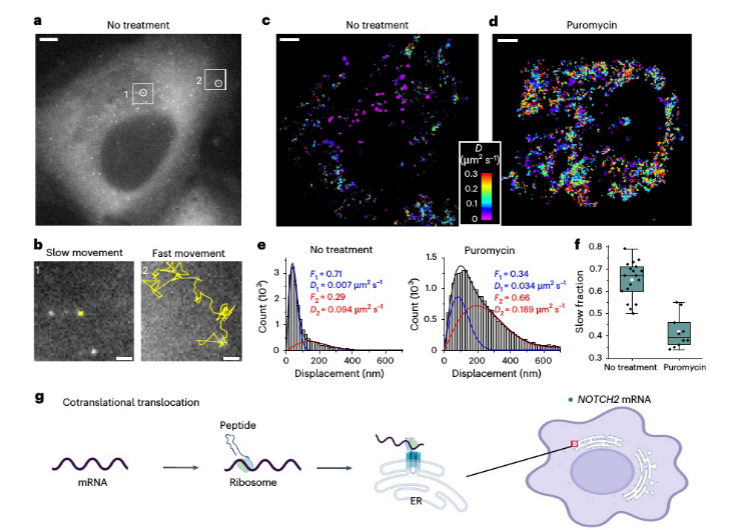
Analysis of the two dynamic populations of NOTCH2 mRNA, the anchoring mechanism of stationary NOTCH2 mRNA on the endoplasmic reticulum, and the changes in dynamic populations after puromycin treatment.
II. Unveiling the Dynamic Mechanisms of mRNA
The research team utilized Single-Molecule Displacement and Diffusion Mapping (SMdM) to thoroughly analyze the dynamic characteristics of NOTCH2 and MAP1B mRNAs. In U2OS cells, NOTCH2 mRNA displayed two distinct dynamic populations: one was almost stationary near the endoplasmic reticulum (ER), while the other moved rapidly. To explore the underlying mechanism of this phenomenon, the researchers used puromycin to inhibit translation elongation. The results showed a significant reduction in the stationary NOTCH2 mRNA population, suggesting that its anchoring to the ER relies on a co-translational translocation mechanism.
In contrast, MAP1B mRNA exhibited clear directional transport, moving along microtubules toward the cell periphery. Through trajectory tracking and kinematic analysis, the researchers observed brief pauses in MAP1B mRNA movement, which they hypothesized could be related to the transient binding and release of molecular motor proteins, such as kinesin-1. Importantly, after puromycin treatment, the movement pattern of MAP1B mRNA did not change significantly, indicating that its transport process is independent of translation.
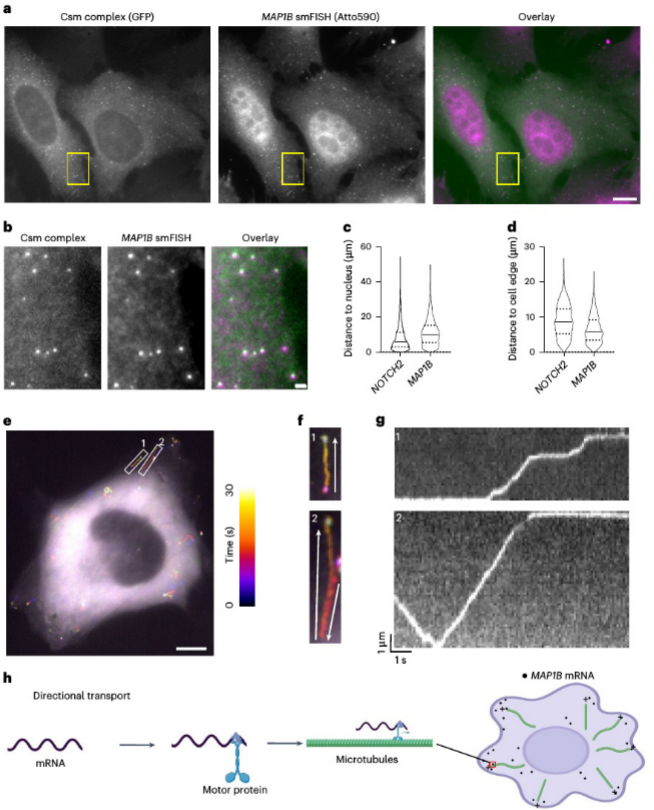
Analyzing the directional transport of MAP1B mRNA, trajectory tracking and kinematic analysis reveal its movement along microtubules toward the cell periphery.
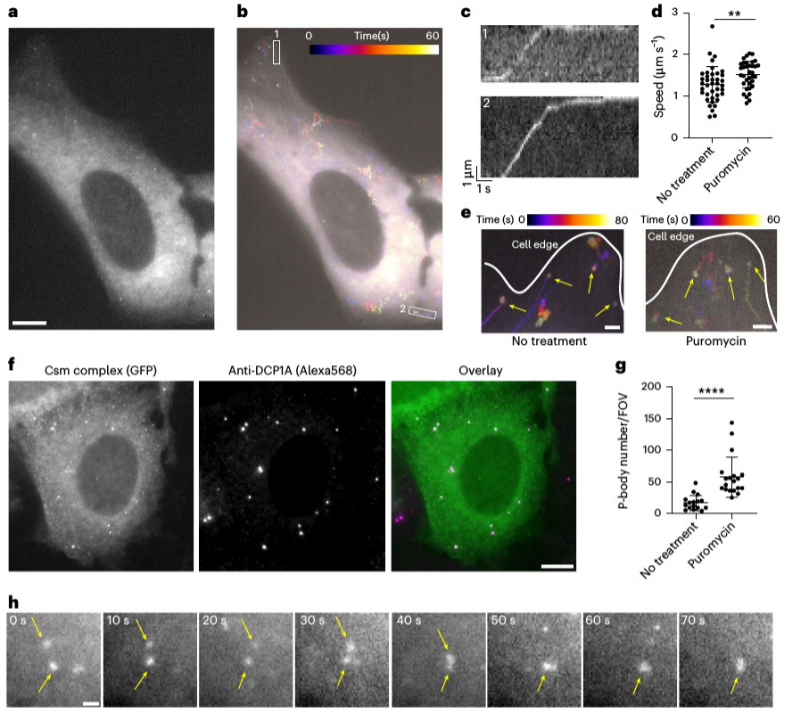
The colocalization experiment of MAP1B mRNA with P-bodies after puromycin treatment demonstrates the role of MAP1B mRNA in the formation of P-bodies.
Furthermore, the research team performed colocalization experiments using the P-body marker DCP1A. They found that after puromycin treatment, some MAP1B mRNA molecules aggregated into larger particles and significantly colocalized with P-bodies, suggesting potential involvement in mRNA degradation or storage processes. Further analysis of single-molecule motion trajectories revealed the microtubule-driven directional transport mechanism of MAP1B mRNA within the cell, and for the first time, real-time visualization of this process was achieved in living cells.
In summary, the smLiveFISH technology introduced in this study successfully overcomes the limitations of traditional RNA imaging methods. It enables tracking the dynamic behavior of unmodified single RNA molecules without altering the stability or translation of the target RNA. More importantly, this technique provides a powerful tool for investigating the spatiotemporal distribution of RNA under both healthy and disease states. CRISPR technology, with its high targeting specificity and programmability, has revolutionized RNA imaging, allowing researchers to precisely target the transcripts of interest for interference-free dynamic monitoring.
Additionally, the use of multiple crRNAs in the CRISPR-Csm system significantly enhances the signal-to-noise ratio, making it possible to reliably detect even low-abundance endogenous RNA. In the future, this technology is expected to expand its applications in neurobiology, cancer research, and other RNA-related diseases, helping scientists unravel the mysteries of RNA dynamics within cells and opening new avenues for RNA-targeted therapies and molecular diagnostics.
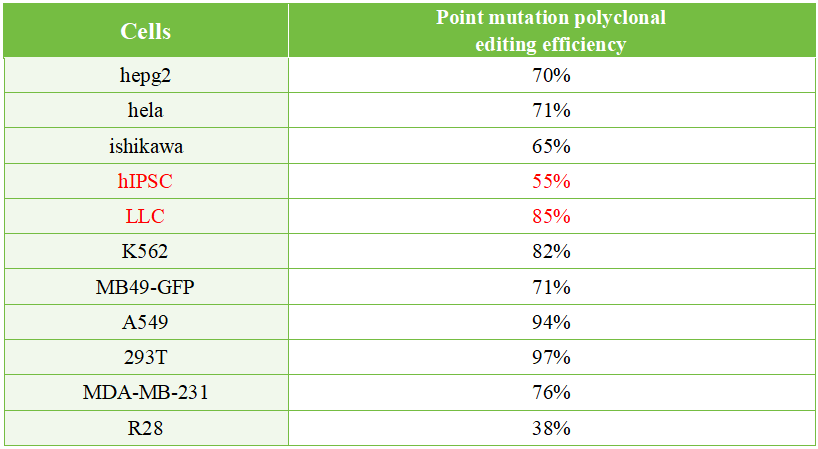
EDITGENE’s BINGO point mutation platform has provided gene point mutation services to thousands of customers. The cell pool editing efficiency reaches up to 95% in cell lines such as 293T and A549, and up to 80% in challenging-to-transfect and edit cell lines like LLC and K562.
Recent Blogs
- 1. [Literature Review] New Developments in Prime Editing: Phage-Assisted Evolution and Protein Engineering Yield More Efficient and Compact Prime Editors
- 2. [Literature Review] A New Hope for Progeria Treatment: CRISPR Gene Editing Uncovers Protein Synthesis Dysregulation and Novel Therapeutic Targets
- 3. [Literature Review] Unveiling T Cell Dynamics: CRISPR Library Screening and Single-Cell Transcriptomics Reveal Gene Regulation Networks
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com















Comment (4)