[Literature Review] Unveiling T Cell Dynamics: CRISPR Library Screening and Single-Cell Transcriptomics Reveal Gene Regulation Networks
CRISPR screening
In January 2025, Professor Alex Marson’s team published a research paper in Nature titled “Central control of dynamic gene circuits governs T cell rest and activation,” with an impact factor of 50.5. The study employed CRISPR screening and single-cell transcriptomics to uncover the dynamic regulatory network governing T cell gene expression in both resting and activated states.
By systematically identifying transcription factors and epigenetic regulators of IL-2Rα expression, the research highlighted the pivotal role of MED12 in T cell state transitions, offering new targets and strategies for immunotherapy.
Original Article Link:https://www.nature.com/articles/s41586-024-08314-y
T cells play a pivotal role in the immune system, and their functional diversity depends on the precise regulation of intracellular gene expression networks. The transition between activated and resting states is crucial for maintaining immune homeostasis. IL-2Rα (CD25) is a key marker of T cell activation, with distinct regulatory patterns of expression among different T cell subsets.
In effector T cells (Teff), IL-2Rα is transiently upregulated following activation; in regulatory T cells (Tregs), however, its expression remains persistently high. The roles of transcription factors and epigenetic regulators in controlling IL-2Rα expression under different cellular conditions remain unclear.
To comprehensively understand the regulatory mechanisms of IL-2Rα, researchers employed CRISPR screening, combined with single-cell transcriptomics and chromatin analyses, to illuminate the dynamic gene expression networks governing T cells in both resting and activated states.
I. CRISPR Screening Process and Results
1. Cell Isolation and Activation
The researchers isolated CD4+ T cells, including regulatory T cells (Tregs) and effector T cells (Teffs), from healthy donor leukopak-derived peripheral blood leukocytes. These cells were then stimulated with CD3/CD28/CD2 activators (ImmunoCult™) to induce activation and IL-2Rα expression. The activated cells were divided into three groups: resting Teff cells, resting Treg cells, and activated Teff cells.
2. sgRNA Library Preparation and Transduction
The researchers used a CRISPR library containing approximately 6,000 single-guide RNAs (sgRNAs) targeting transcription factors and chromatin modifiers expressed in T cells (about 1,350 genes). These sgRNAs were delivered into the activated T cells via viral vectors, introducing a random sgRNA into each cell to facilitate gene knockout.
3. Cell Culture and Screening
After transduction, cells were cultured in IL-2-containing medium to support their survival and proliferation. Screening was performed on the following groups:
Resting Teff cells: IL-2Rα low-expressing cells were screened after 10 days.
Resting Treg cells: IL-2Rα high-expressing cells were screened after 10 days.
Activated Teff cells: Cells were restimulated on day 9, and IL-2Rα high-expressing cells were screened after 72 hours.
4. Cell Sorting and sgRNA Extraction
Using flow cytometry (FACS), cells were sorted into high and low IL-2Rα expression groups. Genomic DNA was then extracted from the sorted cells, and the sgRNA sequences were amplified by PCR and sequenced. The sequencing data were analyzed using MAGeCK software to identify genes that significantly affect IL-2Rα expression under different conditions.
5. Screening Results
Resting Teff cells: Several genes, such as KLF2, SOCS3, and MYB, were identified as negative regulators of IL-2Rα expression in resting Teff cells.
Resting Treg cells: Genes like FOXP3, USP22, and MYC were found to positively regulate IL-2Rα expression in resting Treg cells.
Activated Teff cells: Genes including GATA3, BATF, and IRF4 were identified as positive regulators of IL-2Rα expression in activated Teff cells.
Dynamic Regulation by MED12: Notably, MED12 showed dynamic regulatory roles across different cell types and states. In resting Teff cells, MED12 functioned as a negative regulator of IL-2Rα. Conversely, in activated Teff cells and Tregs, it acted as a positive regulator. Loss of MED12 disrupted T cell state transitions, preventing cells from fully entering either a resting or activated state and instead leaving them in an intermediate state.
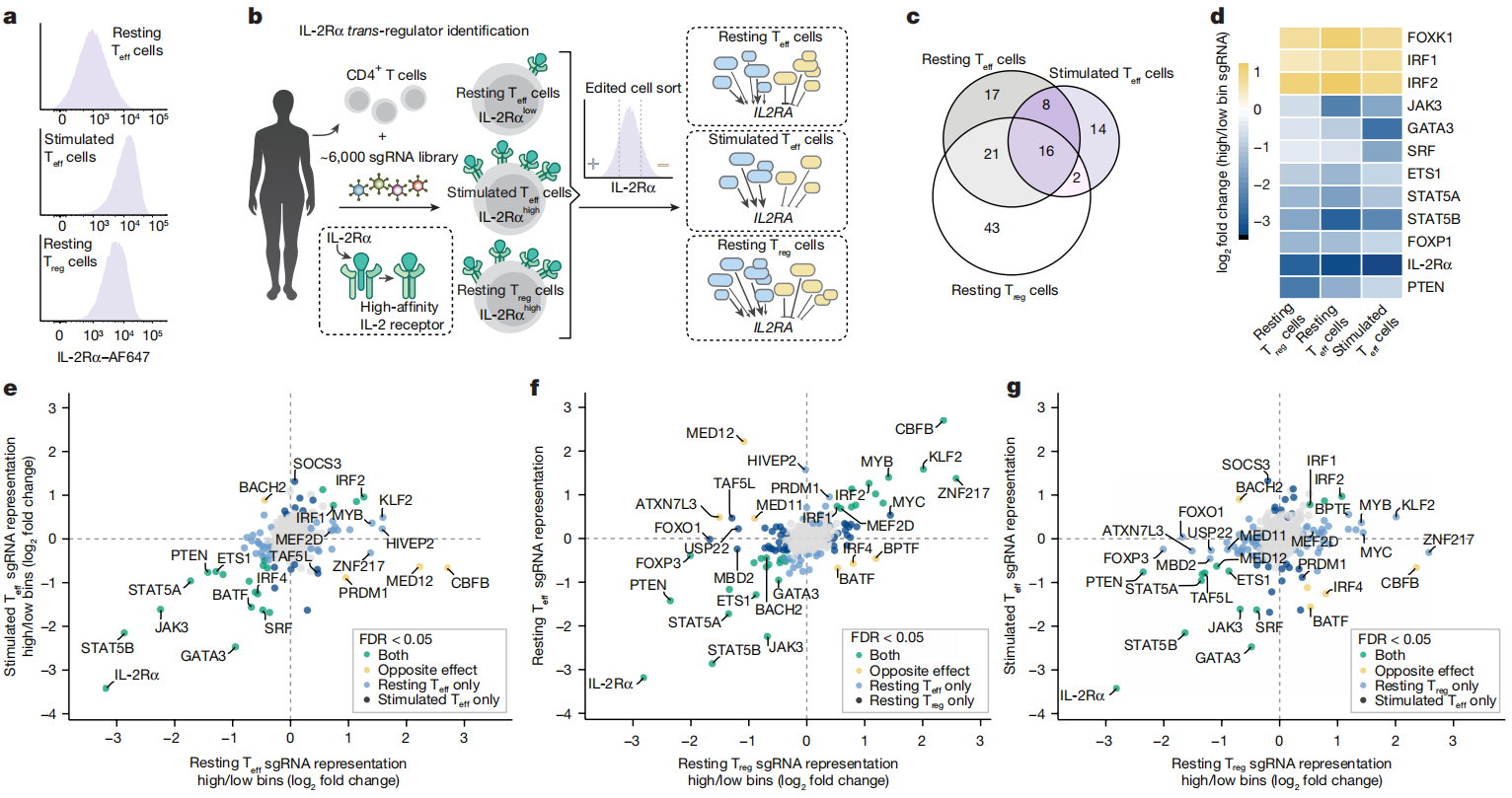
Fig.1 Identification of context-dependent regulators of IL-2Rα expression
II. Single-Cell Transcriptomics Analysis
To further validate the CRISPR screening results and unravel the gene regulatory networks, the researchers performed single-cell transcriptomics (Perturb-seq) analysis on Treg and Teff cells in both resting and activated states. By examining how each gene knockout impacted the global transcriptome, they constructed gene regulatory networks for T cells in different states.
Resting Teff Cells:
KLF2, SOCS3, and MYB were identified as negative regulators of IL-2Rα in resting Teff cells. Knocking out these genes significantly increased IL-2Rα expression.
Activated Teff Cells:
GATA3, BATF, and IRF4 were confirmed as positive regulators of IL-2Rα in activated Teff cells. Knocking out these genes markedly reduced IL-2Rα expression.
MED12’s Global Regulatory Role:
Knocking out MED12 led to increased IL-2Rα expression in resting Teff cells, but decreased expression in activated Teff and Treg cells. This highlights MED12’s critical role in T cell state transitions.
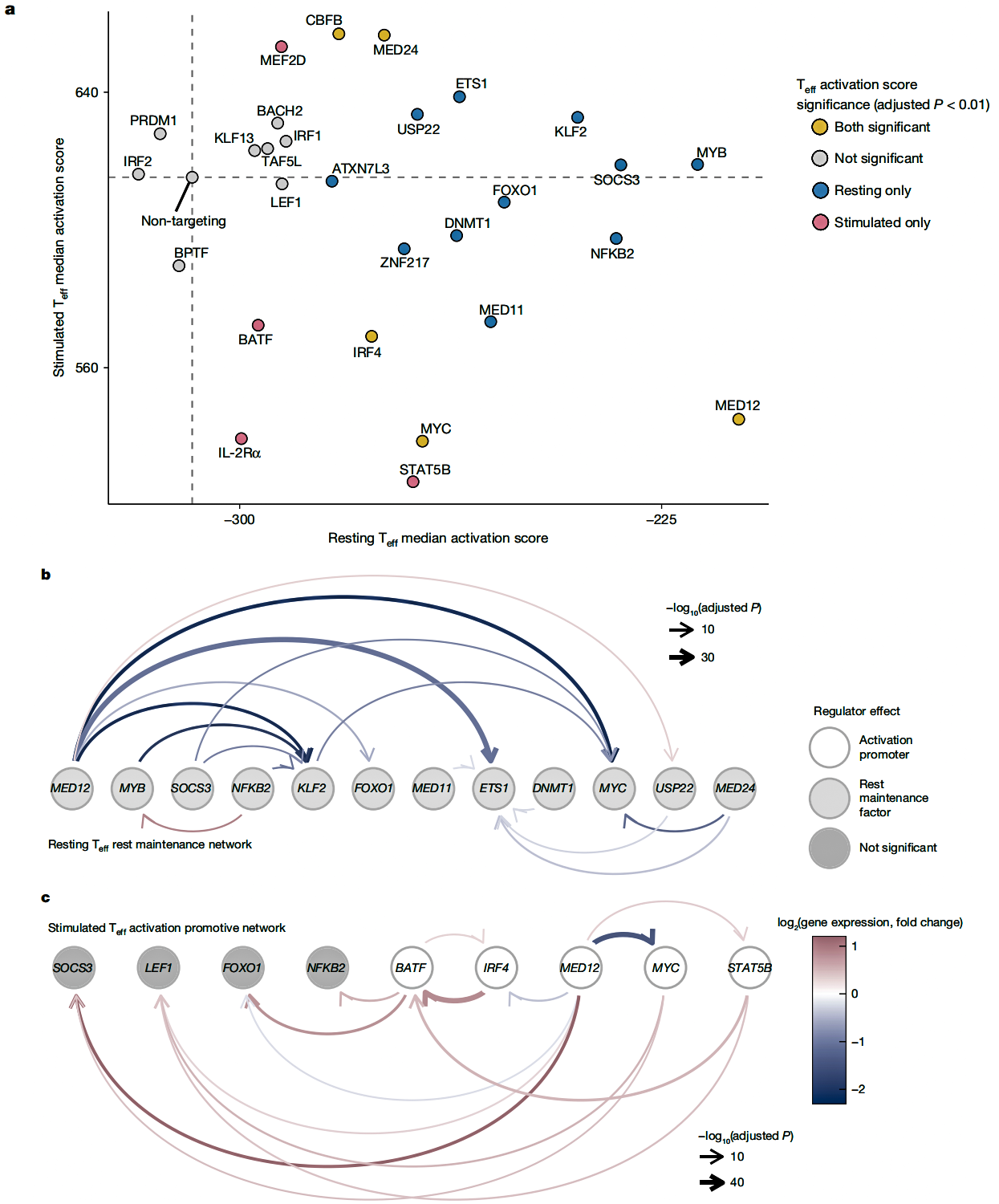
Fig.2 Perturb-seq (single-cell transcriptomics) reveals regulatory networks controlling T cell rest and activation
III. Investigation of MED12’s Regulatory Mechanism
1. Chromatin Analysis
MED12 interacts with the COMPASS complex (a histone methyltransferase complex) to regulate histone H3K4 methylation. In resting Teff cells, the loss of MED12 leads to a reduction in H3K4me3 levels. In contrast, in activated Teff cells, MED12 loss results in increased H3K4me3 levels.
2. Gene Expression Network
MED12 modulates the expression of several key factors (such as KLF2, MYC, and ETS1) to either maintain T cells in a resting state or promote their activation. In resting Teff cells, MED12 upregulates KLF2 expression, which suppresses IL-2Rα. In activated Teff cells, MED12 enhances the expression of MYC and GATA3, thereby promoting IL-2Rα expression.
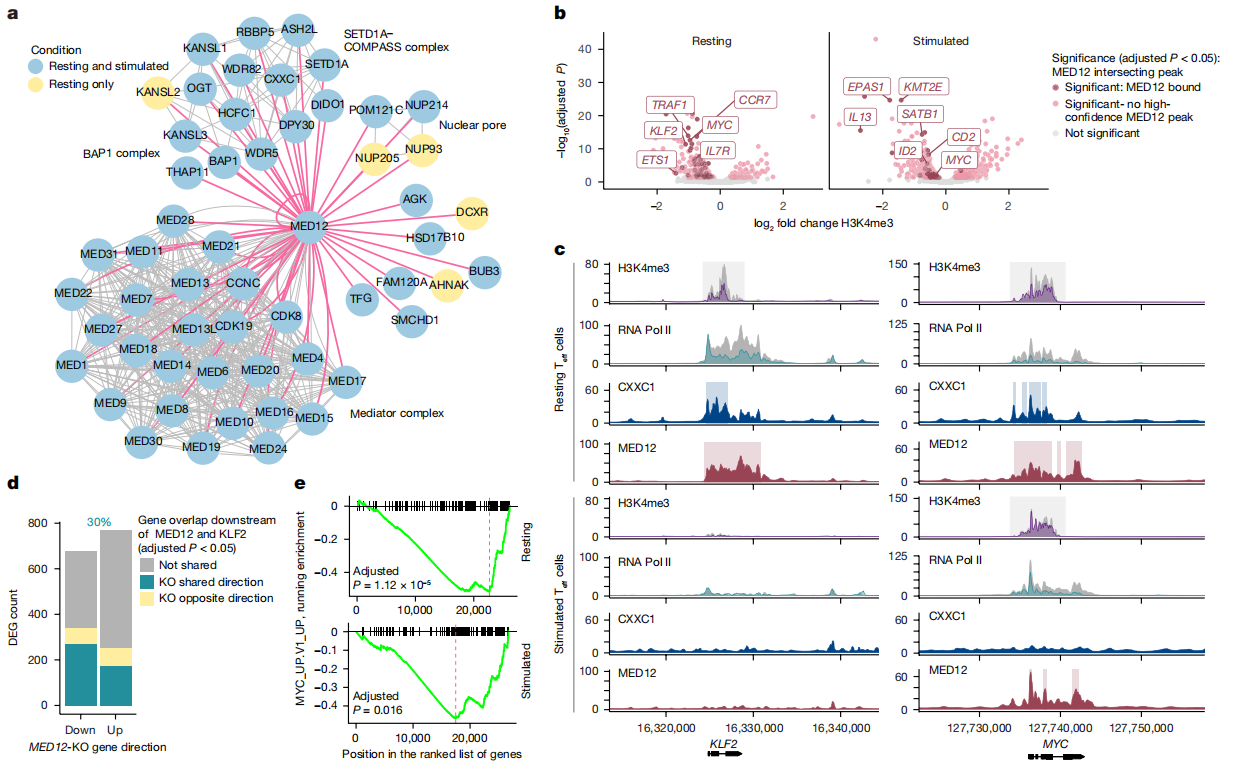
Fig.3 MED12 shapes chromatin to promote cell type-specific and stimulus-specific regulation
IV. Impact of MED12 on Cell States
1. Cell State Transitions
The absence of MED12 prevents T cells from fully transitioning into either a resting or activated state, leaving them in an intermediate state. This underscores MED12’s critical role in T cell state transitions.
2. Cell Survival and Proliferation
Loss of MED12 limits activation-induced cell death (AICD), thereby enhancing T cell survival and proliferation. This could have positive implications for T cell persistence.
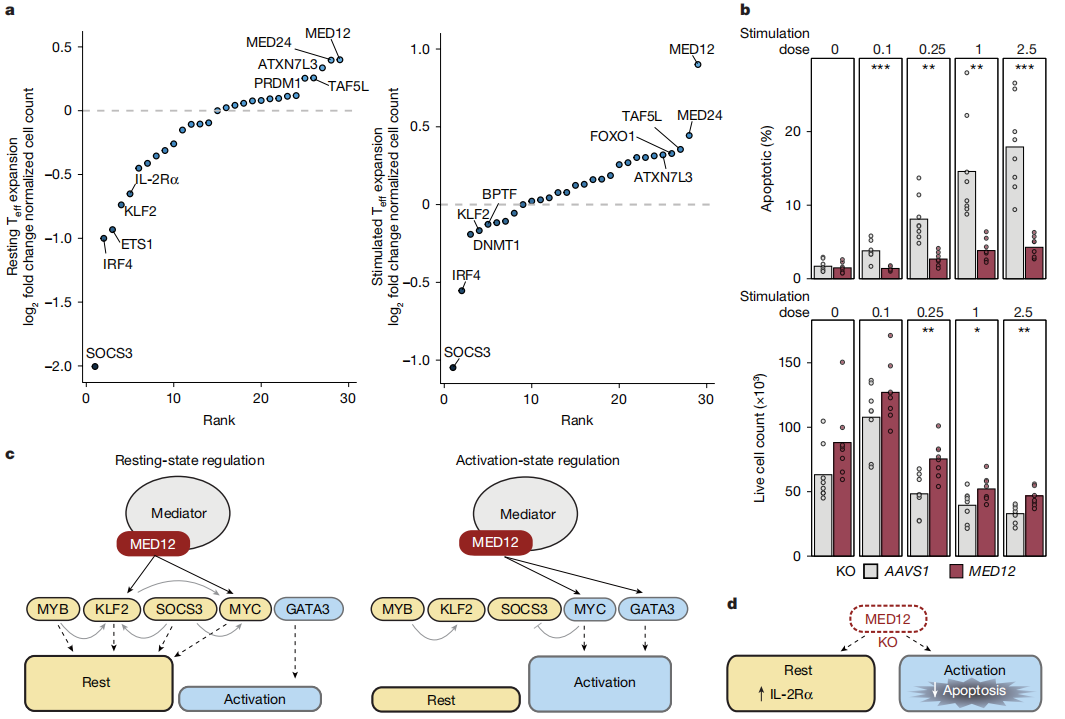
Fig.4 Loss of MED12 limits activation-induced T cell apoptosis
V. Significance of the Study
Through CRISPR screening and single-cell transcriptomics analysis, researchers uncovered the dynamic regulatory networks controlling gene expression in T cells under resting and activated states. The regulatory role of MED12 offers a potential target for developing new immunotherapy strategies. For instance, modulating MED12 activity could enhance T cell persistence and functionality, thereby improving the effectiveness of immunotherapy.
EDITGENE offers a one-stop comprehensive solution for CRISPR library screening, including sgRNA library design and customization, Cas9 stable cell line construction, library lentivirus packaging, library cell generation, functional screening experiments, and NGS analysis. We provide popular libraries such as GeCKO v2 CRISPR and offer the most complete collection of Library Plasmids and library viruses in stock, with delivery within one week. Order now and start screening immediately!
Recent Blogs
- 1. [Weekly News] From "Gene Scissors" to "RNA Erasers": A New Skill of the CRISPR-Cas System Has Been Unveiled
- 2. [Literature Review] Prime Editing Breakthrough: In Vivo Gene Editing Treats Sickle Cell Anemia in Mice
- 3. [Weekly News]: High-Impact Nature Study—CRISPR Library Screening Unveils Synthetic Lethality Potential of PELO Gene in Two Cancer Subtypes
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com
















