[Literature Review] Prime Editing Breakthrough: In Vivo Gene Editing Treats Sickle Cell Anemia in Mice
Prime editing

Prime editing is an innovative gene editing technology that utilizes a fusion of the Cas9 protein and reverse transcriptase to precisely make base substitutions, insertions, or deletions directly within the target DNA—without relying on double-stranded DNA breaks. Compared to traditional CRISPR/Cas9 approaches, prime editing is both more accurate and safer, substantially minimizing the risk of insertions and deletions (indels) and off-target edits.
In February 2023, the journal Blood published a groundbreaking study led by scientists from the University of Washington, Aristotle University of Thessaloniki, and Harvard University. The research team successfully employed in vivo prime editing in hematopoietic stem cells (HSCs) to effectively treat a mouse model of sickle cell disease (SCD). This breakthrough not only brings new hope for the gene therapy of sickle cell disease but also introduces promising new avenues for the treatment of other genetic disorders through advanced gene editing techniques.

Original Article Link:https://doi.org/10.1038/s41586-024-07670-z
Sickle cell anemia is a genetic disease caused by a single-gene mutation in which the sixth amino acid in the β-globin gene (HBB) is changed from glutamic acid to valine. This substitution causes red blood cells to form a sickle shape under low-oxygen conditions, leading to a range of severe pathophysiological effects such as shortened red cell lifespan, vascular blockages, and organ damage. Although current treatments can help alleviate symptoms, they do not cure the disease at its root. Gene therapy, particularly approaches capable of directly repairing the mutated gene, is seen as the key to curing sickle cell anemia.
In this study, the researchers developed a prime editing system based on a Helper-Dependent Adenoviral (HDAd) vector. This vector efficiently delivered the prime editing components to the hematopoietic stem cells of mice, enabling gene editing in vivo.
I. In Vitro Validation: Gene Editing at the Cellular Level
Before applying the Prime Editing system in mice, the research team conducted a series of in vitro validation experiments to ensure the editing tool’s effectiveness and safety.
Cell-Based Experiments: The researchers tested their system on human embryonic kidney cells (HEK293) and chronic myelogenous leukemia cells (K562). The results demonstrated that the Prime Editing system, from PE3 to PE5max, achieved a marked improvement in editing efficiency. The system effectively repaired a target T>A mutation back to the normal GAA sequence, reaching efficiency rates as high as approximately 100% in K562 cells.
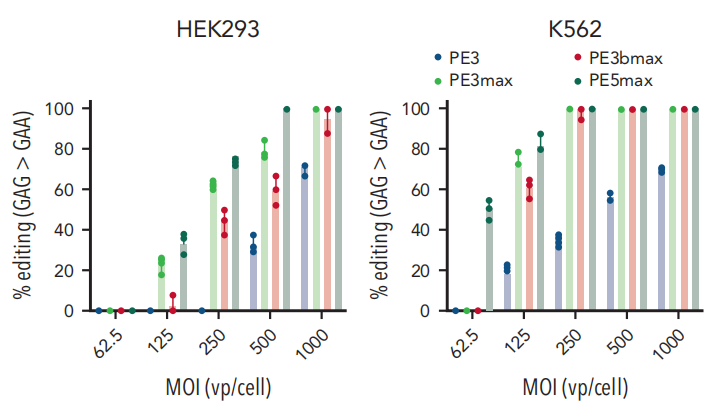
Fig.1 Analysis of G>A (silent PAM site) editing in cell lines
In Vitro Experiments on Patient-Derived Cells : To further evaluate the effectiveness of Prime Editing under pathological conditions, researchers isolated CD34+ hematopoietic stem cells from the blood of patients with sickle cell anemia (SCD) and subjected these cells to Prime Editing treatment in vitro. The results showed a repair rate of 7.5% for the sickle cell mutation in the edited cells. Moreover, edited cells exhibited significant improvements during in vitro differentiation, including reduced oxidative stress markers (ROS) and improved red blood cell morphology. Additionally, only a low level of editing was detected at a single predicted off-target site, which was found to have no functional consequences.
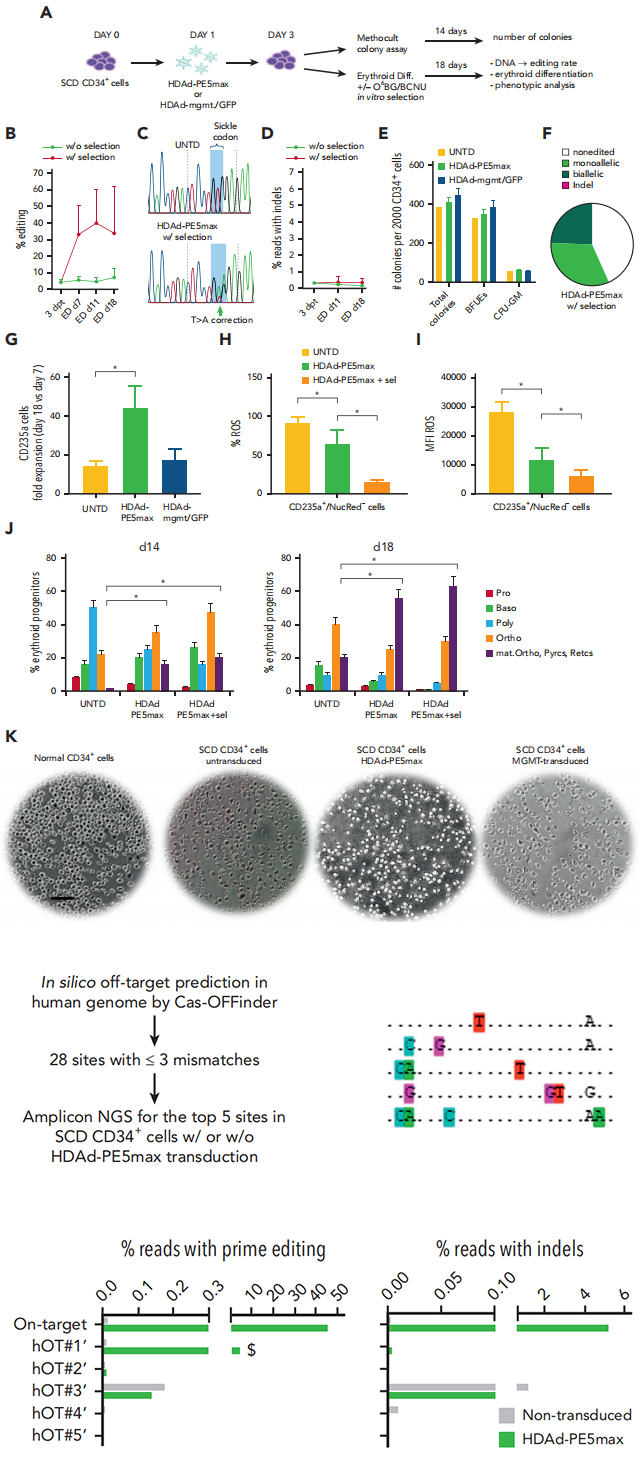
Fig.2 In vitro studies with CD34+ cells from patients with SCD infected with HDAd-PE5max
II. In Vivo Studies: Gene Editing in a Mouse Model
After confirming the effectiveness of the Prime Editing system in vitro, the researchers tested it in a sickle cell anemia mouse model (CD46/Townes mice).
Preparing the Mouse Model: Transgenic mice carrying the human CD46 gene were crossed with Townes mice, which mimic sickle cell disease. This breeding strategy produced CD46/Townes mice, which can efficiently receive the HDAd vector and predominantly express sickle hemoglobin (HbS) in their red blood cells. These mice accurately recapitulate the pathological features of human sickle cell anemia.
In Vivo Gene Editing: The HDAd-PE5max system was administered via intravenous injection. Following the injection, the mice underwent treatment with O6BG/BCNU drugs to select for and expand successfully edited hematopoietic stem cells (HSCs).
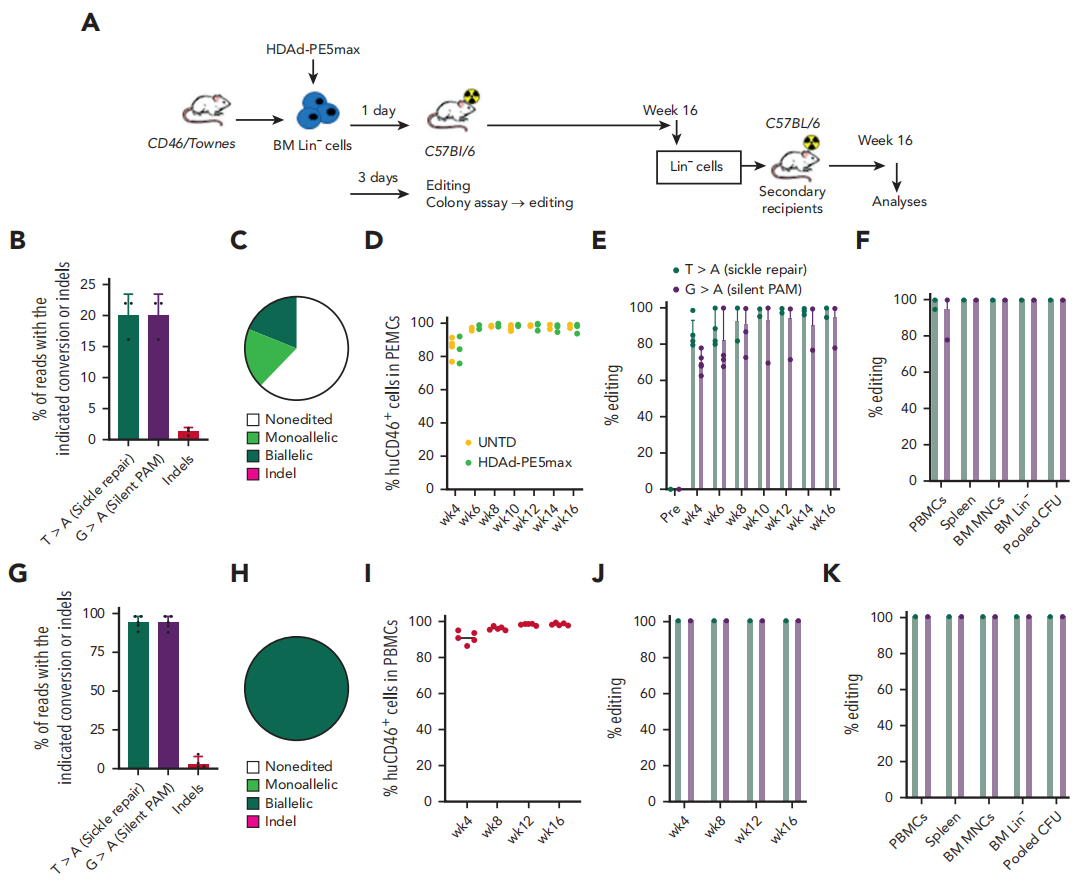
Fig.3 Correction of the SCD mutation by ex vivo HSC transduction
Evaluation of In Vivo Editing Outcomes: Over a 16-week observation period, the research team employed various methods to assess the effects of in vivo gene editing:
Genetic Level: Sanger sequencing and NGS analysis revealed that 43.6% of the sickle cell anemia mutant gene (βS) was corrected to the normal gene (βA) in vivo. Furthermore, the edited HSCs were able to maintain stable gene correction levels in secondary recipient mice, with an average correction rate of approximately 40%.
Protein Level: HPLC and mass spectrometry analysis demonstrated that 43% of the sickle hemoglobin (HbS) in the mice’s blood was replaced by normal adult hemoglobin (HbA).
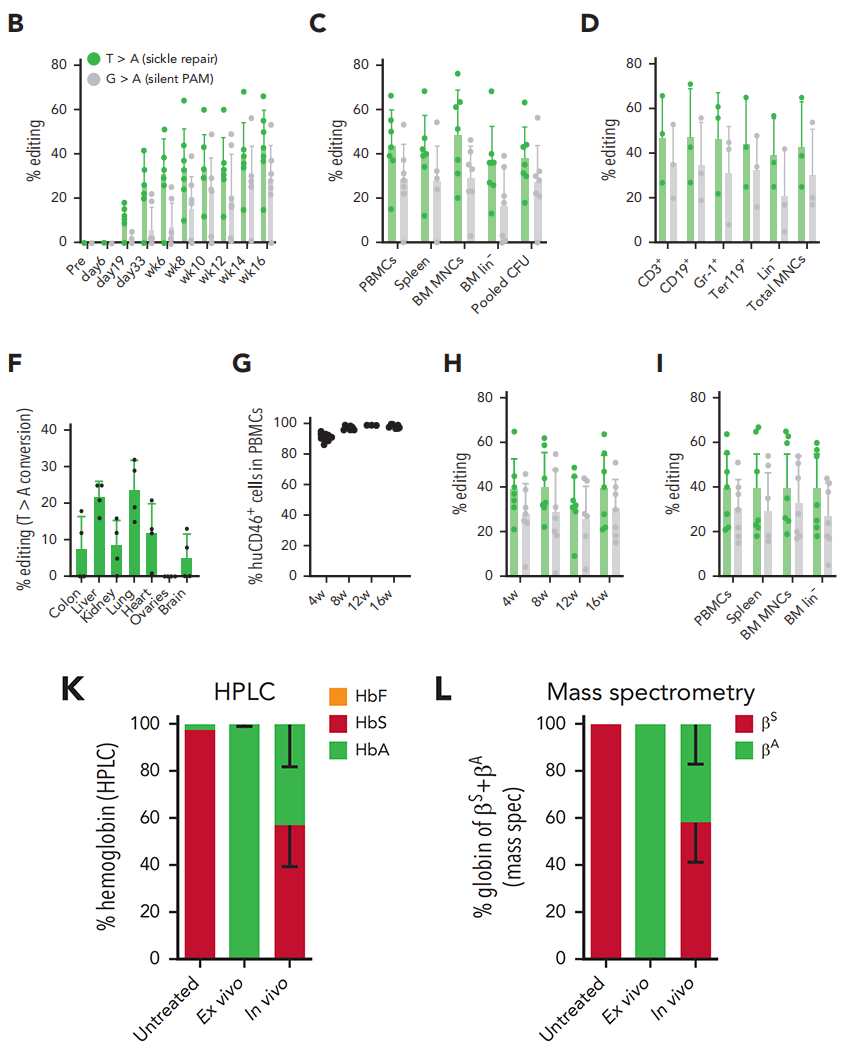
Fig.4 Therapeutic prime editing in CD46/Townes mice after in vivo HSC transduction with HDAd-PE5max
Phenotypic Improvement: Significant improvements were observed in red blood cell morphology, hematological parameters, and histopathological changes. The proportion of sickle cells dropped from 86% to 29.6%.
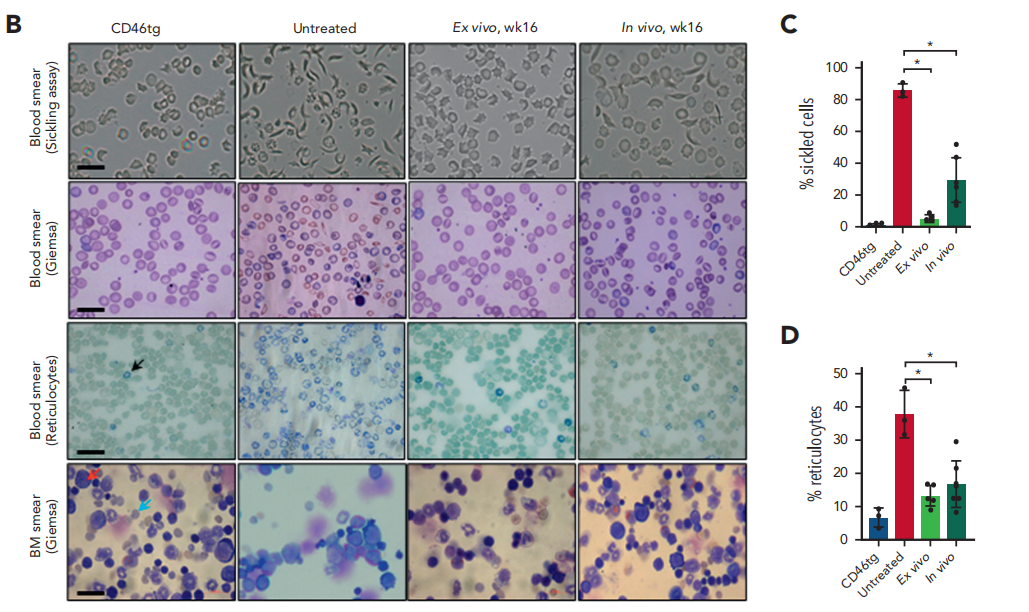 Fig.5 Phenotypic correction of CD46/Townes mice using ex vivo and in vivo approaches at week 16 after treatment
Fig.5 Phenotypic correction of CD46/Townes mice using ex vivo and in vivo approaches at week 16 after treatment
Off-Target Effects Detection: In the mouse model, the Prime Editing system—particularly PE5max—demonstrated extremely high specificity. No significant off-target editing was observed. Even at potential off-target sites predicted by CIRCLE-seq and Cas-OFFinder, editing frequencies were very low and significantly below the editing efficiency at the intended target site.
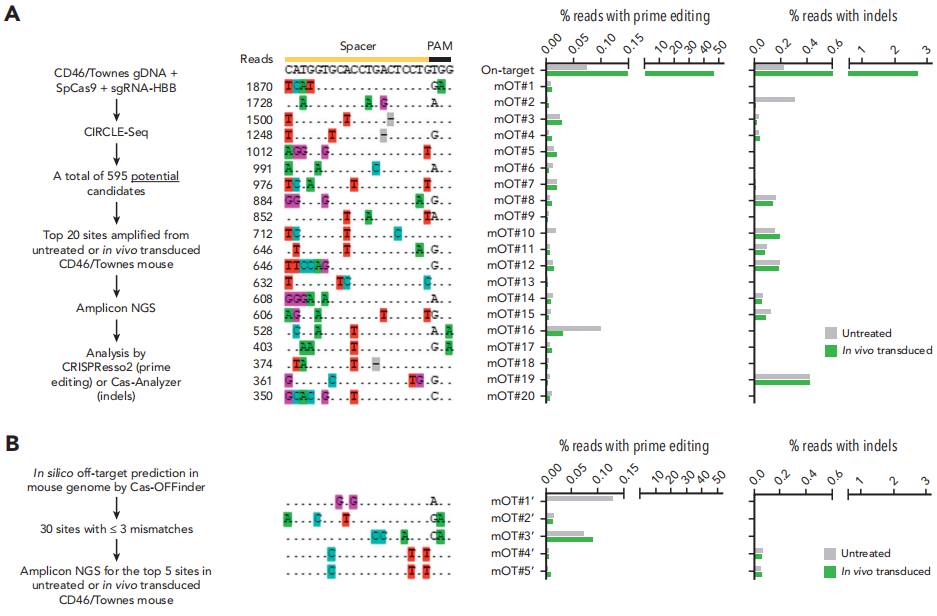
Fig.6 Analyses of OT effects
This study, combining in vitro validation with in vivo application, highlights the enormous potential of Prime Editing in treating sickle cell anemia. From in vitro cell experiments to successful implementation in an in vivo mouse model, these researchers not only demonstrated the efficiency and safety of the Prime Editing system, but also laid a solid foundation for its future clinical applications.
Original Article Link: https://doi.org/10.1182/blood.2022018252
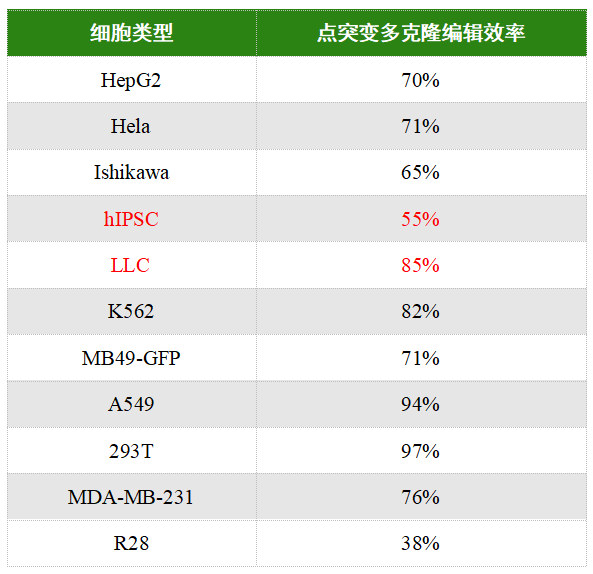
EDITGENE’s BINGO™ point mutation platform has successfully provided gene point mutation services to thousands of clients. With cell pool editing efficiencies reaching up to 95% in 293T and A549 cells—and up to 80% in more challenging cell types such as LLC and K562—this platform achieves success rates far beyond those of traditional gene point mutation methods, delivering precise, highly efficient solutions for research institutions and companies.
Recent Blogs
- 1. [Weekly News] High-Impact Nature Study—CRISPR Library Screening Unveils Synthetic Lethality Potential of PELO Gene in Two Cancer Subtypes
- 2. [Weekly News] New Insights into Type III CRISPR System: Cryo-EM Reveals Selective Synthesis of Cyclic Oligoadenylates (cOA) as a Second Messenger in the System
- 3. [Literature Review] CRISPR Knockout Library Screening Identifies TMEFF1 as a Neuron-Specific Restriction Factor for HSV-1
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com
















