[Research Frontier] CRISPR screen decodes stimulatory responses of primary T cells
CRISPR library screening

CRISPRa libraries by activating specific genes in cells, scientists are able to gain insight into the role of these genes in regulating cellular functions and biological processes. In this article, we introduce a classic application of the CRISPRa library.
A research team at the Institute for Genomic Immunology at Gladstone-UCSF published a study in Science titled CRISPR activation and interference screens decode stimulation responses in primary human T cells . The study reports a genome-wide CRISPR activation (CRISPRa) and interference (CRISPRi) screen of human T cells to identify the gene networks that control interleukin-2 (IL-2) and interferon- γ (IFN- γ ) production. The researchers combined the CRISPRa screen with single-cell RNA sequencing to reveal the mechanisms by which interference regulates T-cell activation and elevates cellular status, as well as genes that reprogram key immune cell functions.

Cytokines regulate T-cell activity in response to stimulation and are essential for balancing the immune response, and cytokine dysregulation may lead to autoimmunity, immunodeficiency, and immune evasion in cancer. Interleukin-2 (IL-2) and interferon- γ (IFN- γ ) are important cytokines that regulate T cells; the former is mainly secreted by CD4+ T cells to allow T cell expansion, which can be applied in autoimmunity and cancer therapy; the latter can be secreted by CD4+ and CD8+ T cells, which is associated with cancer immunotherapy. In this study, researchers developed human T cell CRISPRa and CRISPRi screening platforms to investigate the cytokine production pathways and pathways controlling human T cells.
General CRISPRa screens is limited to small-scale experiments due to the slow delivery system , and higher-speed delivery tools are needed for large-scale screening of genes and pathways that interfere with human T cells. To overcome this limitation, the researchers developed a high-titer lentivirus system using the smallest dCas9-VP64 vector (pZR112), and resulted in an overall transduction efficiency of up to 80% , which effectively enhances the strength of the CRISPRa-SAM system for inducing expression of established surface marker target genes. To validate the practical effectiveness of the system, the researchers performed a genome-wide CRISPRa screen of more than 18,800 protein-coding genes and more than 112,000 sgRNAs, and used flow cytometric fluorescence sorting (FACS) to isolate IL-2-producing CD4+ T cells and IFN-γ-producing CD8+ T cells, which were confirmed by sgRNA quantification that sgRNAs targeting IL-2 and IFN-γ were highly enriched in different cytokines and not significantly enriched in non-targeting controls, the CRISPRa screen was highly reproducible in two different human blood donors, and the statistical analysis of the gene level of the CRISPRa screen for IL-2 and IFN-γ showed 444 and 471 hits, respectively, including 171 shared hits, indicating that the team successfully constructed a reliable large-scale CRISPRa screening platform.
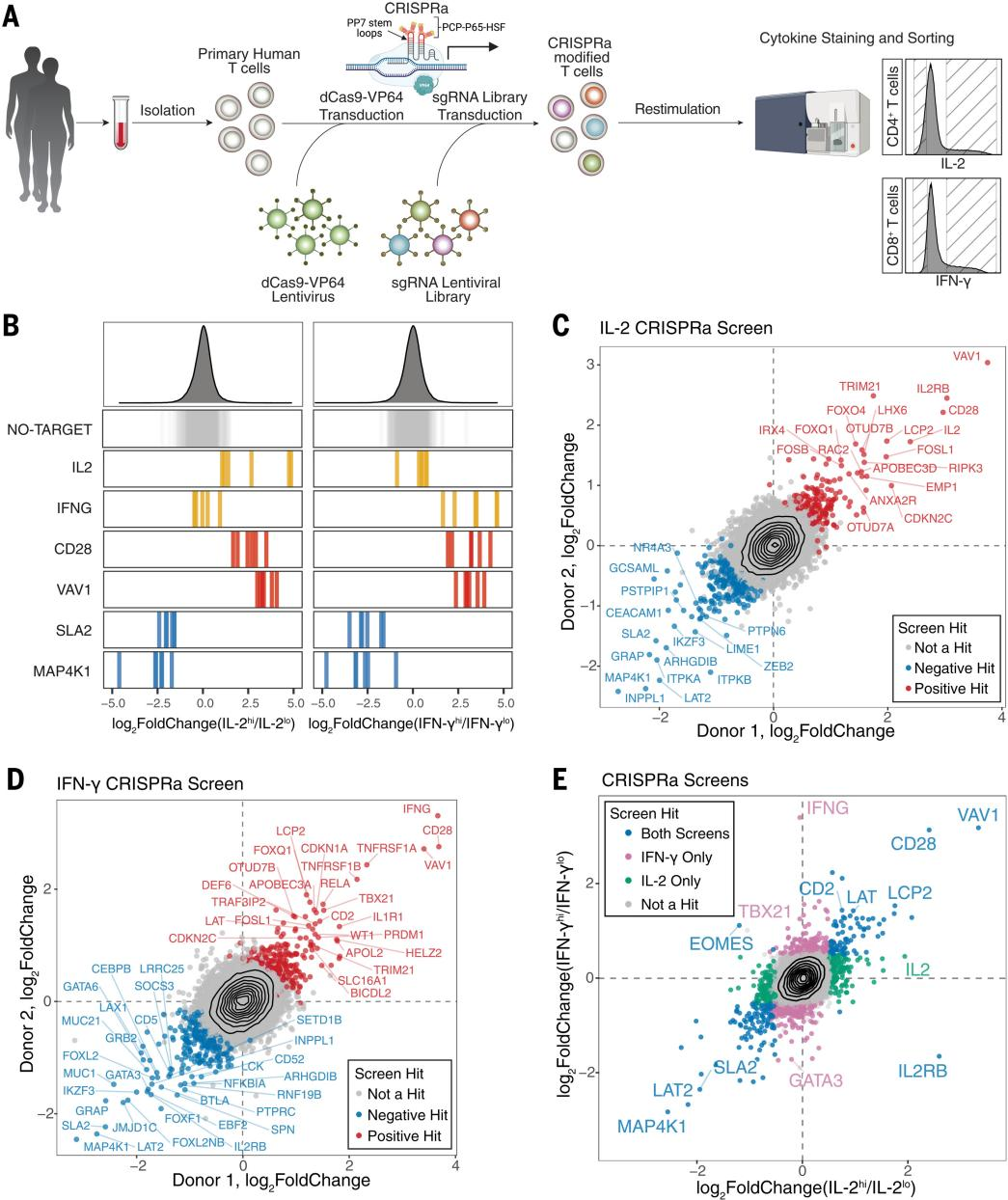
Fig. 1 Genome-wide CRISPRa screen identifies regulators of IL-2 and IFN- γ production in T cells
To study certain cytokines that can only be detected by rendering them dysfunctional, the researchers used CRISPRi and optimized lentiviruses to perform IL-2 and IFN- γ screening experiments, identifying 226 and 203 gene hits, including 92 shared hits. CRISPRi tends to screen for highly mRNA-expressed genes, including the CD3 complex. Whereas CRISPRi failed to detect low or completely unexpressed regulators that were screened for, the researchers used CRISPRa for identification screening and successfully screened for regulators whose expression is only induced in certain T-cell environments, such as PIK3AP1 and IL1R1. Additionally, CRISPRi screened for key components of the Nuclear Factor- к B (NF- к B) pathway, which is required for the production of IFN- γ . T-cell stimulatory signaling promotes IFN- γ production by inhibiting the NF- к B complex (which is encoded by CHUK, IKBKB, and IKBKG) through MALT1, BCL10, TRAF6, and TAK1 (encoded by MAP3K7). CRISPRa, on the other hand, showed a positive set of IFN- γ regulators, including TNFRSF and IL1R1, which also pass through the NF- к B pathway but are not detected by CRISPRi, thus CRISPRa and CRISPRi complement each other to comprehensively discover functional cytokine regulators.
Researchers performed gene enrichment analysis by KEGG to gain insight into functional pathways enriched by CRISPRi and CRISPRa screening. Analysis of data from a large number of genome-wide association studies revealed that CRISPRi and CRISPRa regulators of IFN-γ and CRISPRa regulators of IL-2 are located in regions enriched for immunogenetic traits; integrative analysis of cytokines enriched by the CRISPRa and CRISPRi screens revealed that some of these genes were successfully identified in both screens, suggesting that these genes are core regulators in the stimulus-responsive cytokines produced by T cells; a handful of target genes, including PTPRC (CD45), were affected by both CRISRPa and CRISPRi in the same direction of cytokine production, suggesting that activation and disruption work together to influence the optimal expression levels of certain genes. The researchers used this comprehensive data in conjunction with the literature review to construct a map of the regulatory signaling pathways of cytokine production, including calcium and cytokine signaling pathway genes, and also used CRISPRa to identify regulatory factors that were missing in previous literature, suggesting that complementary screening with CRISPRa and CRISPRi can provide a more accurate and comprehensive map of the regulatory signaling pathways of cytokine production. production and regulatory signaling pathways.
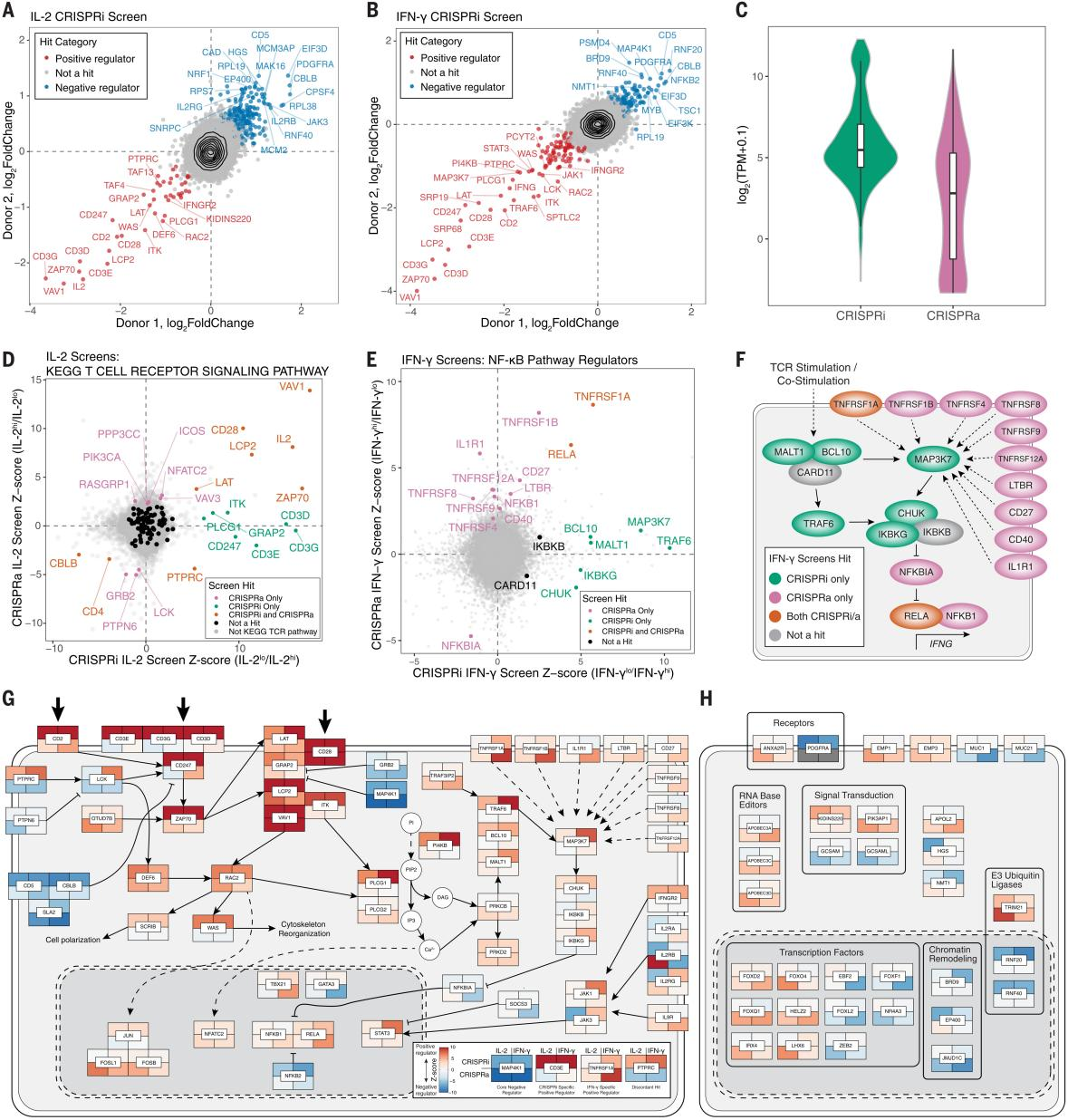
Fig. 2 Complementary CRISPRa and CRISPRi Screens Comprehensively Reveal Cytokine Production Pathways in T Cells
To obtain a deeper phenotypic characterization in the screened hits, the researchers selected 14 different kinds of screened hits for CRISPRa array experiments. The researchers assessed IL-2, IFN-γ, and TNF-α by intracellular staining of CD4+ and CD8+ T cells, and the results showed that 13 of the 14 target genes resulted in significant changes in the proportion of positive cells for the relevant cytokines, and intracellular cytokine staining in the full-array sgRNA group Except for TNFRSF1A, IL2, and IFN-γ, the other regulators in the the absence of stimulation did not spontaneously induce cytokine production. During T-cell differentiation, FOXQ1 and TNFRSF1A significantly reduced the percentage of CD62L+ cells, suggesting a potential mechanism for cytokine transfer to effector T cells. The researchers measured 48 secreted cytokines and chemokines, and after confirming that the effects of the cytokines on IL-2, IFN-γ, and TNF-α were largely consistent with intracellular staining, all cytokines were subjected to major component analysis and hierarchical clustering, with VAV1 and FOXQ1 leading to an increase in the type 1 signature cytokines and suppression of the type 2 cytokines; and OTUD7B as the positive regulator of proximal TCR signaling significantly increased type 2 cytokines. The researchers performed bulk RNA sequencing (RNA-seq) of FOXQ1 and control sgRNA CD4+ T cells and found a strong correlation with secretosome effects, identifying regulators that not only regulate TCR stimulation and signaling, but also regulate T-cell secretomes.
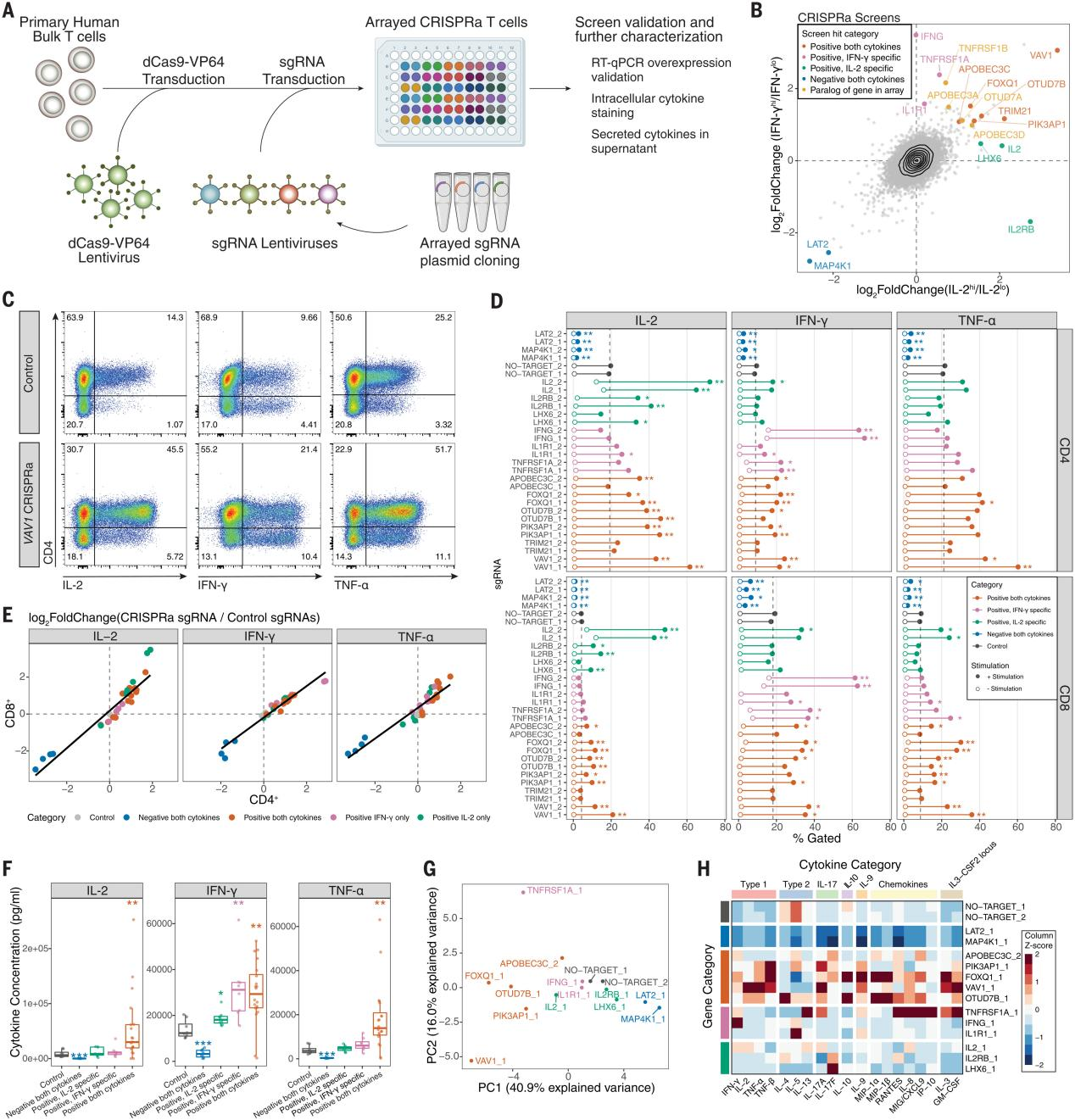
Fig. 3 Characterization of CRISPRa screening hits by array analysis
The researchers developed the CRISPRa Perturb-seq platform for assessing the overall molecular signature generated by each CRISPRa gene induction, and analyzed stimulus-responsive regulators in approximately 56,000 primary human T cells, confirming that sgRNAs can lead to a significant increase in the expression of their target genes. The researchers examined the activation scores of CRISPRa perturbations, and with the exception of IKZF3 (encoding the Aiolos transcription factor), the negative regulators reduced the activation scores, suggesting that they can broadly attenuate the intensity of the stimulus, whereas IKZF3 reduced the expression of IFN- γ but did not lower the overall activation scores, suggesting that there may be a different mechanism of cytokine gene regulation.
The researchers used cluster analysis to characterize the cellular state of CRISPRa-driven cells in both secondarily stimulated and resting T cells, identifying the most up-regulated gene expression markers and cytokine genes, revealing the diversity of CRISPRa-promoted T cell states. The researchers also identified two distinct cell clusters (clusters 1 and 12) expressing IFN-γ. Cluster 1 was labeled for high expression of CCL3 and CCL4 and was enriched for sgRNAs; cluster 12 was enriched for sgRNAs that activate the NF-кB pathway.These results suggest that augmented stimulation/co-stimulation may drive T cells into an activated IFN-γ-expressing state and that a TNFRSF receptor gene subpopulation activation also promoted the cellular state. Thus, CRISPRa Perturb-seq reveals how cytokine-produced regulators modulate T-cell activation and move cells into different stimulus-responsive states.
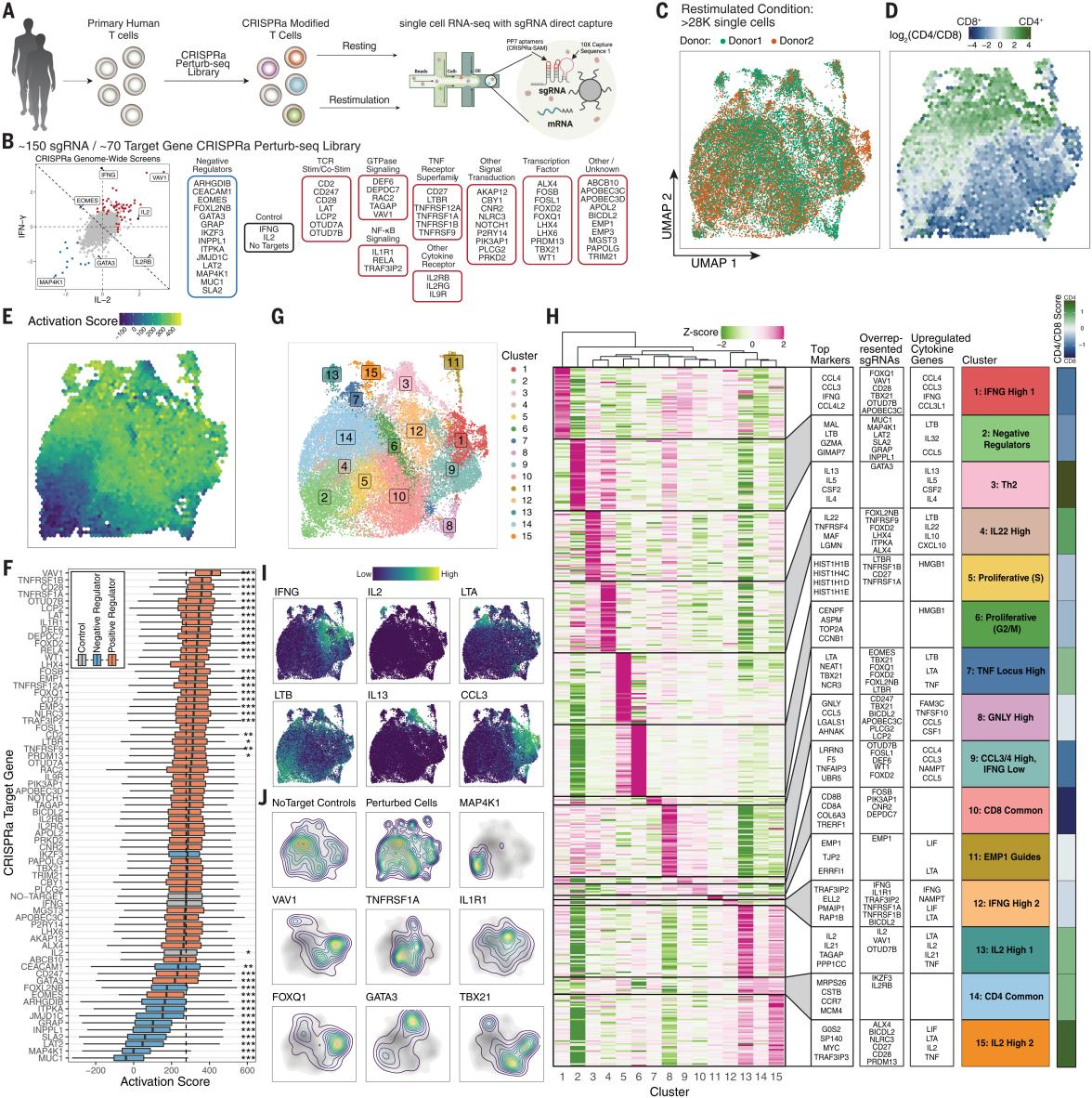
Fig. 4 CRISPRa Perturb-seq captures distinct T-cell states driven by genome-wide cytokine screening
In summary, the researchers have identified required cytokine regulators through CRISPRi, revealed key signaling bottlenecks in pathway function through CRISPRa, and regulators that are not necessarily active in in vitro cultured T cells. The technology developed in this study will enable methods to screen human primary T cells and potentially other primary cell types, and the T cell CRISPRa and CRISPRi screening platforms will contribute to the study of cytokine production pathways and pathways that control human T cells.
EDITGENE provides a complete solution for CRISPR library screening, including library design, customization, lentivirus packaging, positive/negative screening , etc. 90+ CRISPR libraries, plasmids and lentivirus are available! Click to browse our list.
Recent posts
[DEC Bestselling Products] GLP1R, CTNNB1, PKM Knockout Cell Lines
[Research Frontier] New Trends in Prime Editing: Point Mutated Cells for Preclinical Gene Therapy
Prime editing 101 : what is prime editing?
Contact Us
Email:info@editxor.com
Phone:833-2263234 (USA)















Comment (4)