[Star of the Month] Human Genome KO Library,Mouse Kinome CRISPR KO Library,Yeo Lab RNA-binding Protein CRISPR KO Library

CRISPR Library Screening is a high-throughput gene screening method based on CRISPR/Cas9 technology. It involves constructing a library containing thousands of sgRNAs, cloning these sgRNAs into lentiviral vectors, and infecting target cells at a low MOI. This approach ensures that each cell is infected by only one sgRNA, allowing for functional screening of genes corresponding to different sgRNAs. We recommend three best-selling gene libraries from E DITGENE in stock, along with related research literature to aid in your scientific endeavors.
I. CRISPR Knockout Library Screening Identifies Genes Involved in EGF-Induced Apoptosis
Original Link: https://doi.org/10.4143/crt.2022.1414
The expression of Epidermal Growth Factor Receptor (EGFR) is associated with various cancers. Existing EGFR inhibitors, such as tyrosine kinase inhibitors or monoclonal antibodies, have limited efficacy and might induce cancer resistance. Therefore, there is a need to develop innovative drugs targeting EGFR-overexpressing cancer cells.
The mechanisms of EGF (Epidermal Growth Factor)-induced apoptosis are not yet fully understood. To investigate whether the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway and the mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK/ERK) pathway might be involved in this apoptosis, researchers use CRISPR KO library screening techniques to understand EGF-induced apoptosis at the genomic level and explore its potential as a therapeutic target.
Researchers utilized a human genome-scale CRISPR knockout library (GeCKO library v2) containing 123,411 single-guide RNAs (sgRNAs) to knockout 19,050 genes. They analyzed the read counts of each sgRNA obtained from next-generation sequencing using the edgeR algorithm and examined the relationship between EGFR mutation status and the most relevant resistance genes through the cBioPortal for Cancer Genomics database.
The study identified 266 genes potentially responsible for EGF-induced apoptosis and discovered that DDX20, LHFP, REPS1, DUSP1, and KRTAP10-12 are major EGF resistance genes. Among these, DUSP1 appears to be a key component of EGF-induced apoptosis and a novel therapeutic target for EGFR-overexpressing cancers.
Through screening with the Human GeCKO Lentiviral sgRNA Library v2 , it was found that DUSP1 plays a crucial role in EGF-induced apoptosis and may become a novel therapeutic target for EGFR-overexpressing cancers, which has significant implications for developing new cancer treatment strategies.
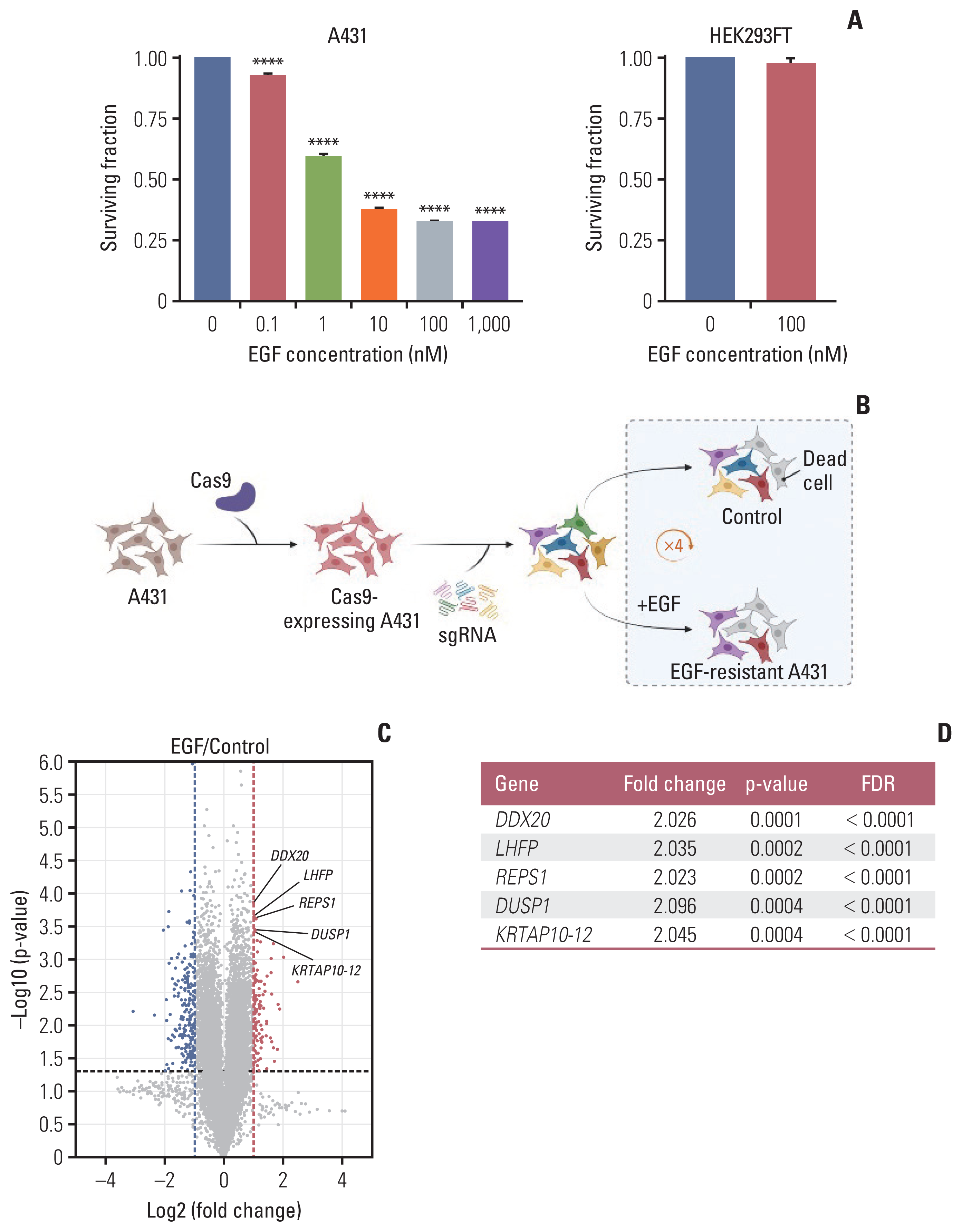
Figure 1 CRISPR/Cas9 Knockout Screening Using A431 Cells
II. Mouse Kinase Library Screening Aids in Discovering New Radiosensitization Targets for Pancreatic Cancer
Original Link: https://doi.org/10.3390/cancers14020326
Pancreatic cancer, particularly pancreatic ductal adenocarcinoma (PDAC), is one of the leading causes of cancer-related deaths worldwide. Conventional treatment methods have limited efficacy in treating PDAC, with surgical resection benefiting only a small subset of patients, chemotherapy (CT) having severe side effects and being ineffective for non-resectable tumors, and radiation therapy resistance complicating treatment.
Therefore, there is a need to identify new therapeutic strategies to address the shortcomings of current treatments. CRISPR-Cas9 screening technology, as a powerful gene-editing tool, can be utilized to discover sensitive targets for pancreatic cancer radiotherapy, and to enhance treatment outcomes by developing and applying targeted inhibitors in combination with radiation therapy.
In this study, researchers utilized CRISPR-Cas9 technology in combination with the Mouse Brie kinome pooled library (a mouse kinase sgRNA library) to perform a genome-wide screen in pancreatic cancer cells. They cultured and screened pancreatic cancer cell lines, followed by functional studies on pancreatic cancer-related genes, including gene knockout and validation as well as drug screening.
The results ultimately identified DYRK1A as a novel therapeutic target for overcoming radiotherapy resistance in pancreatic cancer. Knockout or inhibition of DYRK1A significantly enhanced the sensitivity of pancreatic cancer cells to radiotherapy. Additionally, the combination of Harmine and radiotherapy further increased the radiosensitivity of pancreatic cancer cells.
Researchers utilized the Mouse Kinome CRISPR Knockout Library (Brie) for screening, which not only revealed the role of DYRK1A in radiotherapy resistance in pancreatic cancer but also provided new directions for future clinical research and treatment. This discovery holds significant importance for developing potential strategies for treating pancreatic cancer.
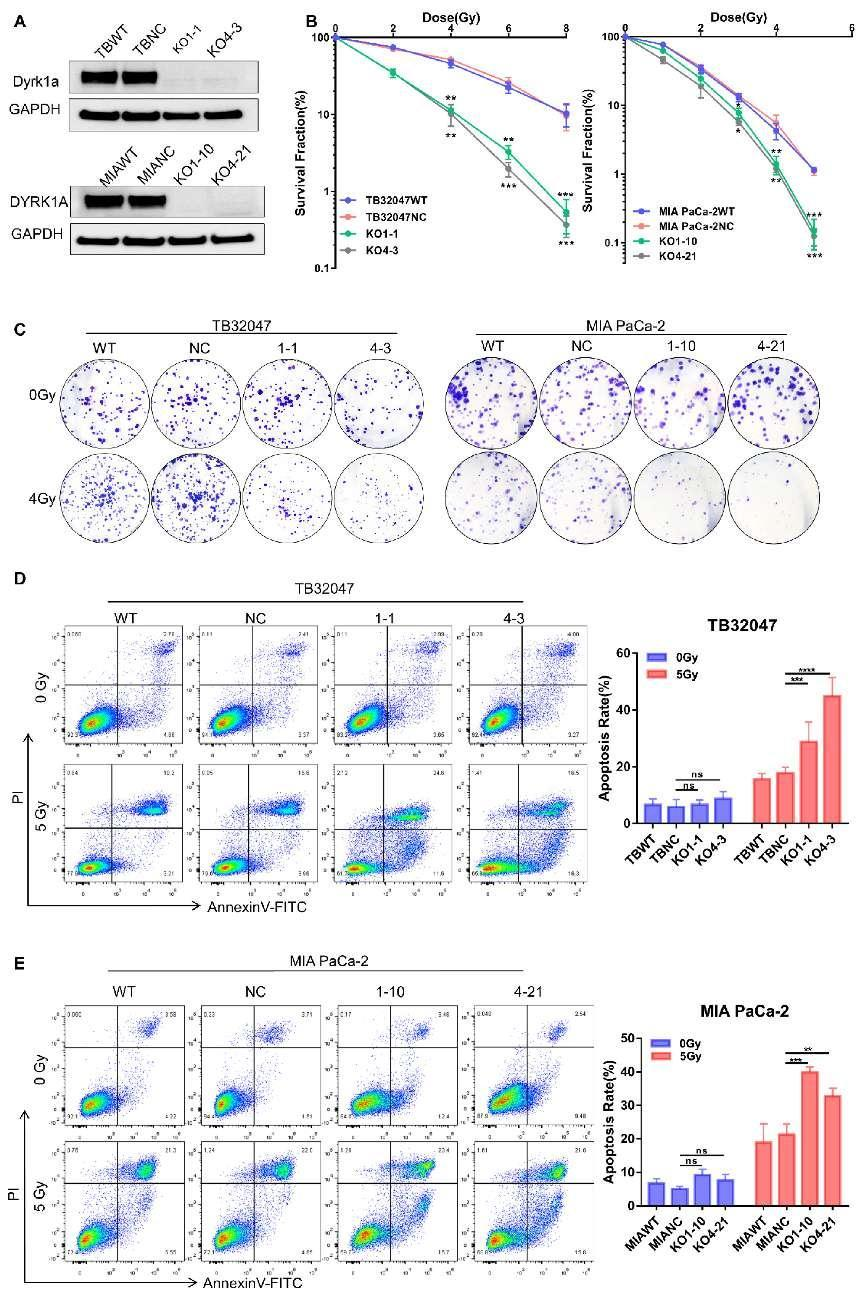
Figure 2 DYRK1A Knockout Enhances Radiotherapy Efficacy in Pancreatic Cancer
III. RNA-Binding Protein Library Identifies INTS3 as an Anti-Apoptotic RNA-Binding Protein and Therapeutic Target for Colorectal Cancer
Original Link: https://doi.org/10.1016/j.isci.2024.109676
Colorectal cancer (CRC) is one of the cancer types with high incidence and mortality rates. Despite improvements in surgical resection, adjuvant chemotherapy, and targeted therapy, many patients still face issues such as unresectable tumors, tumor recurrence, or metastasis. Identifying new therapeutic targets is an urgent need to improve therapeutic efficacy of CRC.
The abnormal expression of RNA-binding proteins (RBPs) can promote tumorigenesis and is considered a potential therapeutic target. Therefore, many researchers aim to use CRISPR-Cas9 technology to comprehensively study RBPs, hoping to gain a deeper understanding of their specific roles in CRC development, thereby providing more comprehensive information for developing targeted therapies.
In this study, researchers used the CRISPR RNA-binding protein (RBP) library to investigate RBPs critical for CRC transmission and survival. They identified INTS3 as an RBP that is highly expressed in CRC and associated with poor clinical prognosis. Subsequently, through RNA sequencing, in vivo experiments, and other series of experiments, they revealed the crucial role of INTS3 in CRC. They also validated that nanoparticle-mediated targeted therapy against INTS3 is an effective strategy for treating colorectal cancer, providing new perspectives and methods for CRC treatment.
Overall, researchers utilized the Yeo Lab RNA-Binding Protein Pooled CRISPR Knockout Library to identify INTS3 as a key RBP closely associated with CRC. They demonstrated a novel therapeutic strategy, offering new approaches and methodologies for CRC treatment. This research holds significant scientific importance and potential clinical application prospects.
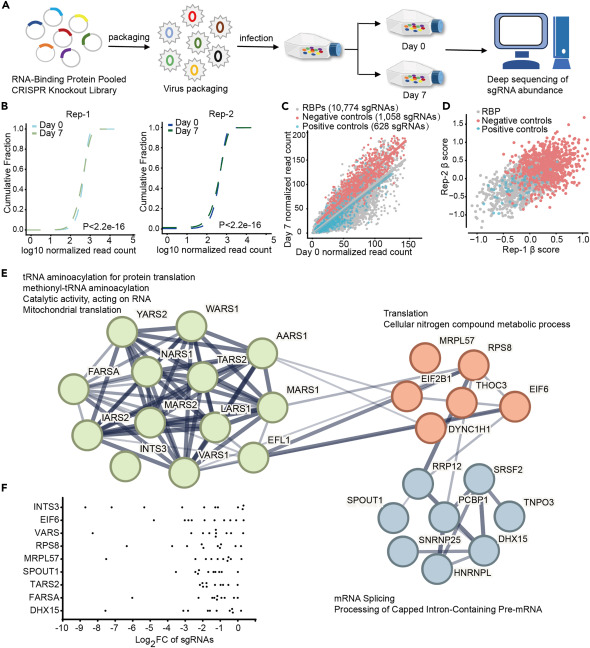
Figure 3 CRISPR-Cas9 screening of RBPs in CRC cells
EDITGENE , specializing in gene editing for over a decade, assembled the most popular types of CRISPR sgRNA library types in the research community, with a coverage rate of ≥99% and uniformity <10. In-stock items are available within weeks, with screening starting immediately upon ordering!
Recent Blogs:
1.【Literature Review 】 The CRISPRi Screen Helps Find Key Genes that Regulate Glioblastoma
3.【Cutting-Edge News】New Trends in Gene Editing - FGFR1, BRD4, and BACE1 Knockout Cells
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com















![[Literature Review] A Novel Mechanism of Cisplatin Resistance in Osteosarcoma: CRISPR Screening Identifies Key Regulators in Organoid Models](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)
![[Quality Share] Decoding Point Mutations: A Comprehensive Guide to Three Common Construction Methods](/uploads/20250328/ESzk5OC49wpxIHVv_3cbfa5e98ea1d238127fe23c72b0f4b2.png)

Comment (4)