[Literature Review] CRISPR screening reveals that removal of TREX1 enhances CRISPR-Cas9-mediated homology-directed repair

CRISPR-Cas9-mediated homology-directed repair (HDR) is a critical technology for introducing precise mutations at targeted genomic loci. However, achieving high HDR efficiency remains a challenge, particularly in cell types with insufficient DNA repair activity. In most human cells, HDR introduces exogenous DNA sequences during the repair of CRISPR-Cas9-mediated double-strand breaks (DSBs), but its efficiency is significantly lower compared to non-homologous end joining (NHEJ). Factors such as the cell cycle stage, differential gene expression, and cellular context can all influence HDR efficiency. Furthermore, patient-derived cells with mutations in DNA repair genes, such as those from individuals with Fanconi anemia (FA), exhibit reduced HDR efficiency, complicating the gene editing process.
To identify the factors that limit CRISPR-Cas9-mediated homologous recombination (HDR), particularly in specific cell types such as those from Fanconi anemia (FA) patients, researchers from the Swiss Federal Institute of Technology Zurich and the Memorial Sloan Kettering Cancer Center conducted a series of experiments. They performed whole-genome CRISPR screening in lymphoblastoid cell lines derived from FA patients, resulting in the publication of their findings, titled "Removal of TREX1 activity enhances CRISPR-Cas9-mediated homologous recombination", in Nature Biotechnology (IF: 33.1). The researchers discovered that the exonuclease TREX1 significantly impacts HDR efficiency; knocking out TREX1 or using chemical protection on DNA templates can reactivate HDR efficiency in TREX1-expressing cells. This study proposes strategies to enhance CRISPR-Cas9-mediated HDR, particularly in contexts of high TREX1 expression.

Original Article Link:https://doi.org/10.1038/s41587-024-02356-3
I. Removal of TREX1 reactivates HDR in FA patient cells
This study utilized a BFP-to-GFP reporter system to assess the HDR efficiency of CRISPR-Cas9-mediated gene editing in FANCA-/- and FANCD2-/- lymphoblastoid cell lines (LCLs) derived from patients. The research induced the conversion from BFP-His151 to GFP-Tyr151 using Cas9 ribonucleoprotein (RNPs) and single-stranded oligodeoxynucleotide (ssODN) templates. Initial experiments indicated that the HDR efficiency in FA-LCLs was significantly lower than that of wild-type cells, particularly in FANCA-/- cells (e.g., FANCA-/- 0.3±0.2%, FANCD2-/- 2.3±0.6%, WT 9.1±0.7%).
To elucidate the factors regulating HDR in FANCA-/- cells, the researchers conducted a whole-genome CRISPR screening using CRISPR interference (CRISPRi) and CRISPR RNPs, combined with fluorescence-activated cell sorting (FACS) to convert BFP to GFP and sequence the recovered sgRNAs. Following lentiviral engineering and electroporation, approximately 0.5% of GFP+ cells were isolated. Next-generation sequencing (NGS) results revealed that the TREX1 gene was significantly enriched in cells with high HDR activity (FDR < 1 × 10-7), indicating that TREX1 is a key regulatory factor for HDR in FANCA-/- cells.
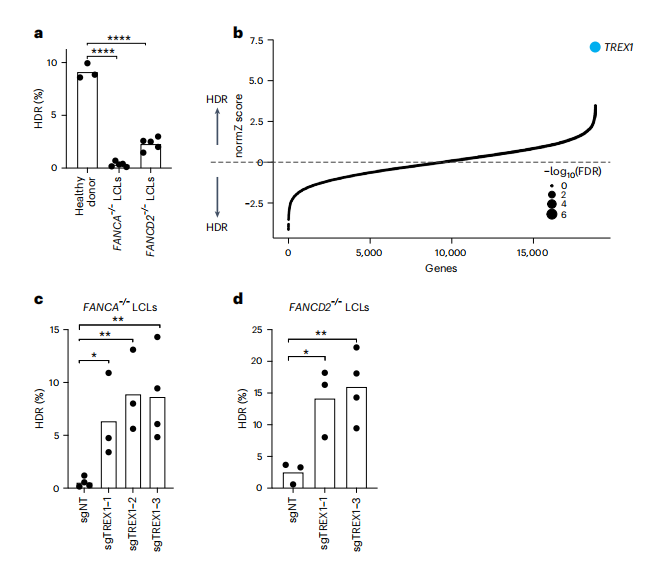
Fig.1 Identification of TREX1 as a restrictive factor of CRISPR-Cas9-mediated HDR
II. TREX1 destabilizes unprotected HDR templates
TREX1 is a 3′-5′ exonuclease localized on the outer membrane of the endoplasmic reticulum (ER) and plays a critical role in suppressing the innate immune response to cytoplasmic DNA by negatively regulating the chronic activation pathway of cyclic GMP-AMP synthase (cGAS). TREX1 primarily degrades single-stranded and double-stranded DNA, and mutations in the TREX1 gene are associated with autoimmune diseases such as Aicardi-Goutières syndrome. In this study, TREX1 was identified as a key factor limiting CRISPR-Cas9-mediated HDR in FANCA-/- cells. Knocking out TREX1 in FANCA-/- and FANCD2-/- cells reactivated HDR, while the overexpression of wild-type TREX1 nearly completely abolished HDR efficiency. Subsequent experiments confirmed that TREX1 interacts with single-stranded oligodeoxynucleotides (ssODNs) and degrades cytoplasmic DNA templates prior to nuclear diffusion, thereby directly reducing HDR efficiency. CRISPR-Cas9 editing conducted in HeLa and RPE-1 hTERT cells demonstrated the activity of this exonuclease, revealing that protecting the 3' end of ssODNs significantly enhances HDR efficiency across various cell types, underscoring the role of TREX1 in limiting template stability during the gene editing process. Despite the strong association between TREX1 and ssODNs, it does not translocate to the nucleus following DNA damage, indicating that its primary role in restricting HDR occurs in the cytoplasm.
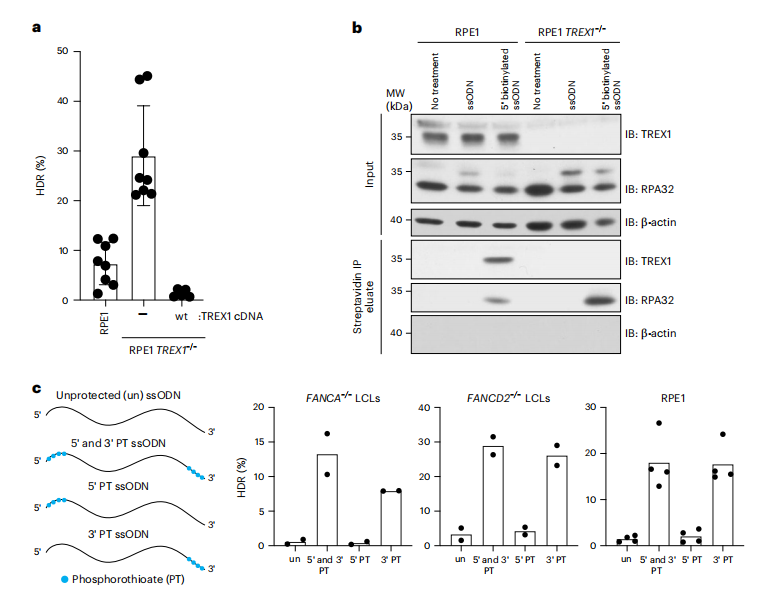
Fig.2 TREX1 interacts with ssODN HDR templates
III. TREX1 restricts HDR efficacy in multiple cell backgrounds and is circumvented by template protection
The study investigated the influence of TREX1 on CRISPR-Cas9-mediated homology-directed repair (HDR) across various human cell types, revealing that cell lines with high TREX1 expression, such as HeLa, exhibited lower HDR efficiency. In contrast, cells with low TREX1 expression, such as K562 and HEK293, demonstrated higher HDR levels. By employing CRISPR interference (CRISPRi) to knock down TREX1 or utilizing protected single-stranded oligodeoxynucleotides (ssODNs), the researchers were able to enhance HDR efficiency in TREX1-expressing cells.
The findings were further extended to primary human cells, showing that the use of protected ssODNs significantly increased HDR rates in hematopoietic stem cells (HSPCs) and activated T cells, while having minimal effects on induced pluripotent stem cells (iPSCs). Moreover, knocking out TREX1 in T cells resulted in augmented HDR from unprotected ssODN templates, achieving efficiency comparable to that observed with protected ssODN templates. These results indicate that TREX1 serves as a crucial regulator of HDR, and that strategies such as ssODN protection or TREX1 inhibition can improve CRISPR-Cas9 editing efficiency across different cell types. Furthermore, the study suggests the potential for combining TREX1 inhibition with other HDR-enhancing techniques, such as the DNA-PKcs inhibitor AZD7648, to further enhance gene editing outcomes.
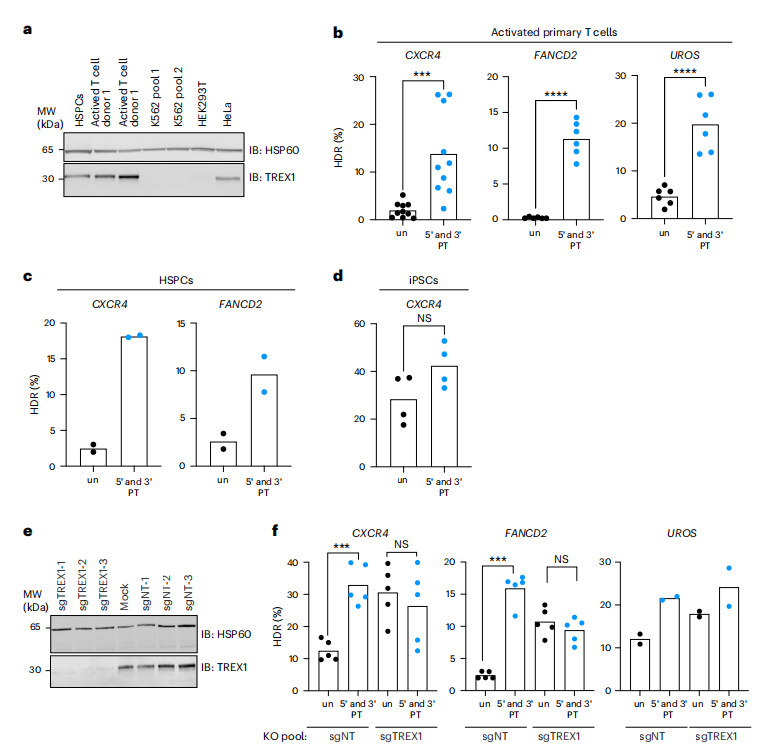
Fig.3 TREX1 suppresses HDR in TREX1-expressing primary cells
IV. HDR donor type affects TREX1 activity
The study broadened its investigation into the impact of TREX1 on various types of DNA donors used in genome editing, moving beyond single-stranded oligodeoxynucleotide (ssODN) templates. It assessed the influence of TREX1 on circular and linear double-stranded DNA (dsDNA) as well as recombinant adeno-associated virus (rAAV) donors, which are often utilized for the insertion of large DNA sequences. Through gene editing experiments with plasmid dsDNA donors in both TREX1-suppressed and wild-type HeLa cells, the researchers found that HDR efficiency was not significantly affected by TREX1 suppression in the presence of covalently closed plasmid templates.
In contrast, the use of linear PCR-amplified dsDNA donors revealed that TREX1 suppression significantly enhanced HDR efficiency in both HeLa and Jurkat cells, suggesting that TREX1 expression negatively impacts HDR when DNA donors possess free 3′ ends. Further supporting this notion, the overexpression of TREX1 in K562 cells dramatically reduced the efficiency of HDR from an ssODN template. The researchers also examined the role of TREX1 in rAAV donor-mediated HDR and discovered that, while TREX1 overexpression diminished HDR efficiency with ssODN templates, its effect was less pronounced with AAV donors. This discrepancy is attributed to AAV donors' capacity to deliver DNA cargo into the nucleus, theoretically avoiding interaction with ER-localized TREX1. Nonetheless, extended exposure to AAV donors was found to reduce HDR in TREX1-expressing cells. Collectively, these findings strongly indicate that TREX1 is a key regulator of CRISPR-Cas9-mediated HDR, particularly for linear, unprotected DNA donor molecules, and that chemically protecting the 3′ ends of DNA templates can effectively circumvent TREX1 activity.
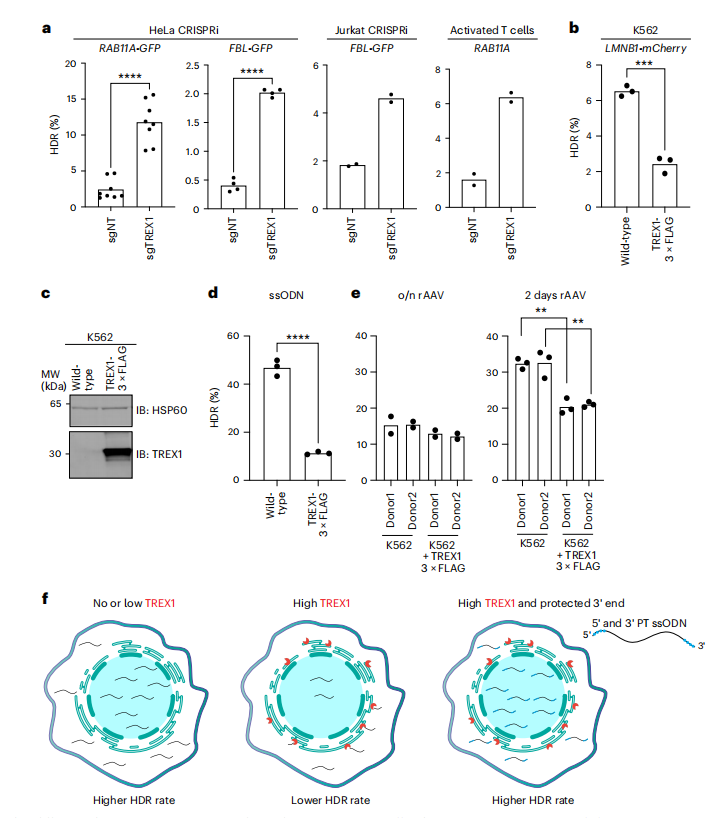
Fig.4 TREX1 has differential activity on various types of HDR donors
In summary, this study found that TREX1, as an exonuclease, affects the efficiency of CRISPR-Cas9-mediated homology-directed repair (HDR) in human cells, particularly in those with elevated TREX1 levels, such as Fanconi Anemia (FA) cells and various immortalized cell lines. To address this issue, removing TREX1 or using chemically protected single-stranded DNA templates can significantly enhance HDR efficiency in cells with high TREX1 expression. The study indicates that protecting the ends of HDR templates can shield them from the action of TREX1, thereby improving both HDR efficacy and the overall efficiency of CRISPR gene editing. Additionally, it underscores the importance of TREX1 as a biomarker for template protection, highlighting its substantial potential in advancing CRISPR therapeutic strategies.
EDITGENE provides one-stop solution for CRISPR library screening, CRISPR KO/CRISPRa/CRISPRi all are available, and the most complete library plasmid/virus in stock in the market, in-stock delivery in one week, order now, screen instantly!
Recent Blogs:
2.[Weekly News] A Novel type VII CRISPR-Cas system: Discover the Potential of CRISPR Gene Editing
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com










![[Literature Review] A “Gene Scalpel” Emerges: A New Hope for Tay-Sachs Disease Cure](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)