[Literature Review] Unlocking CRISPR-Cas12a's Full Potential: Enhancing Trans-Cleavage with Variants
CRISPR-Cas12a

CRISPR-Cas12a has gained widespread use in molecular diagnostics due to its efficient trans-cleavage DNA nuclease activity. While Cas12a's trans-cleavage ability enables signal amplification for diagnostic applications, its sensitivity is still insufficient for clinical use without pre-amplification steps.
Therefore, enhancing Cas12a's trans-cleavage activity is necessary to improve the detection limit for CRISPR-Cas12a-based diagnostics. Studies have shown that the single-strand ends generated by cis-cleavage products can competitively bind to the catalytic site, sterically hindering the binding of reporter DNA and trans-cleavage.
Based on this, a team of researchers from South Korea has proposed a strategy to enhance Cas12a's trans-cleavage activity by reducing steric inhibition from cis-cleavage products. Their findings were recently published in Biosensors and Bioelectronics, under the title Enhanced trans-cleavage activity using CRISPR-Cas12a variant designed to reduce steric inhibition by cis-cleavage products.
Original Article Link:https://doi.org/10.1016/j.bios.2024.116859
I. Removing steric hindrance from cis-cleavage products enhances trans-cleavage activity.
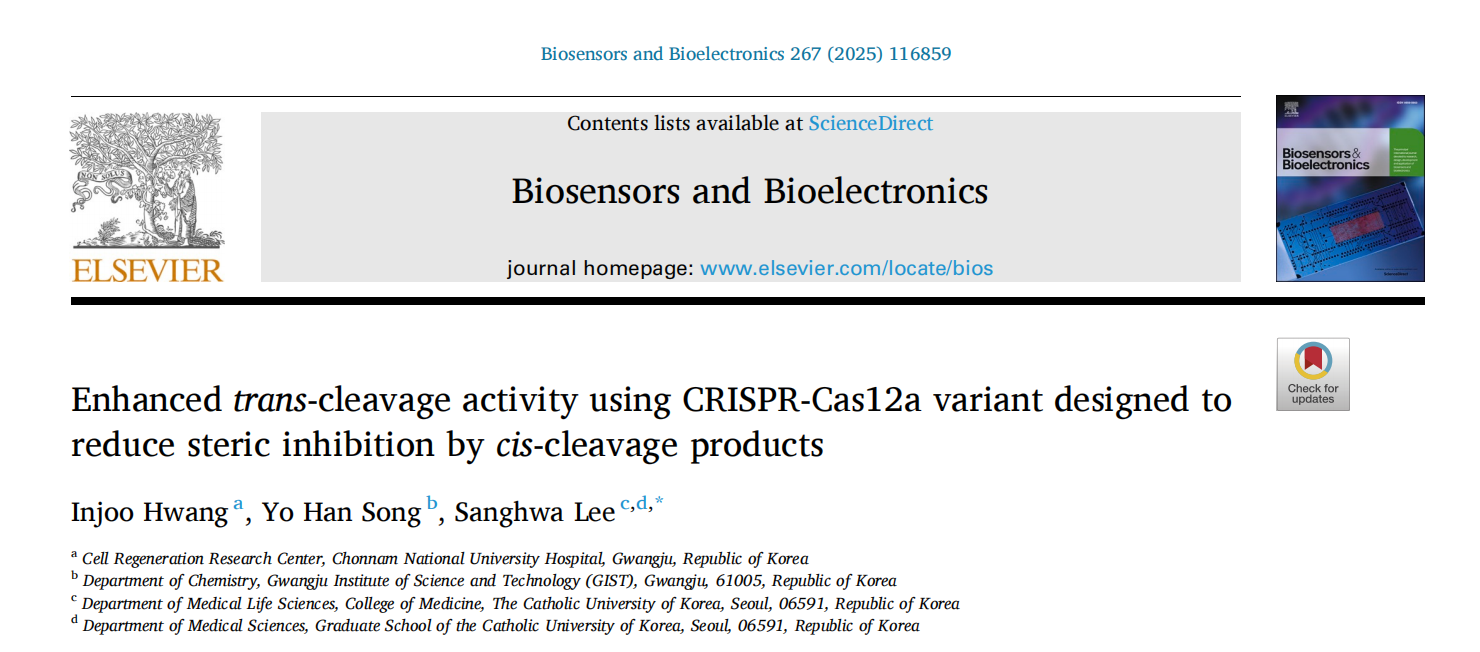
Previous research has shown that pre-removal of double-strand cleavage ends significantly boosts Cas12a's trans-cleavage efficiency. Similar results were observed in fluorescent trans-cleavage experiments. In these experiments, researchers used DNA activators with target strand (TS) protruding ends of varying lengths to measure trans-cleavage activity. The time trajectories were fitted using a zero-order reaction model to obtain the apparent trans-cleavage rates (kobs) for different DNA activators.
The results revealed that for a 19-nt TS activator, the apparent trans-cleavage rate was 3.69 times higher than that of the full-length TS activator, indicating that removing the TS protruding end can significantly enhance Cas12a’s trans-cleavage activity.
To further investigate whether TS protruding ends sterically hinder the loading of trans-DNA substrate in the trans-cleavage reaction, the researchers examined the trans-cleavage activity of W355A Cas12a and the effect of pre-removal of TS protruding ends on its activity. The W355A mutant, which employs faster DNA unwinding and subsequent TS loading, accelerated TS cleavage.
The results showed a significant reduction in trans-cleavage activity with the W355A mutant, consistent with the hypothesis that TS protruding ends increase binding to the catalytic site of W355A Cas12a, thereby sterically hindering the binding of the trans-cleavage DNA substrate.
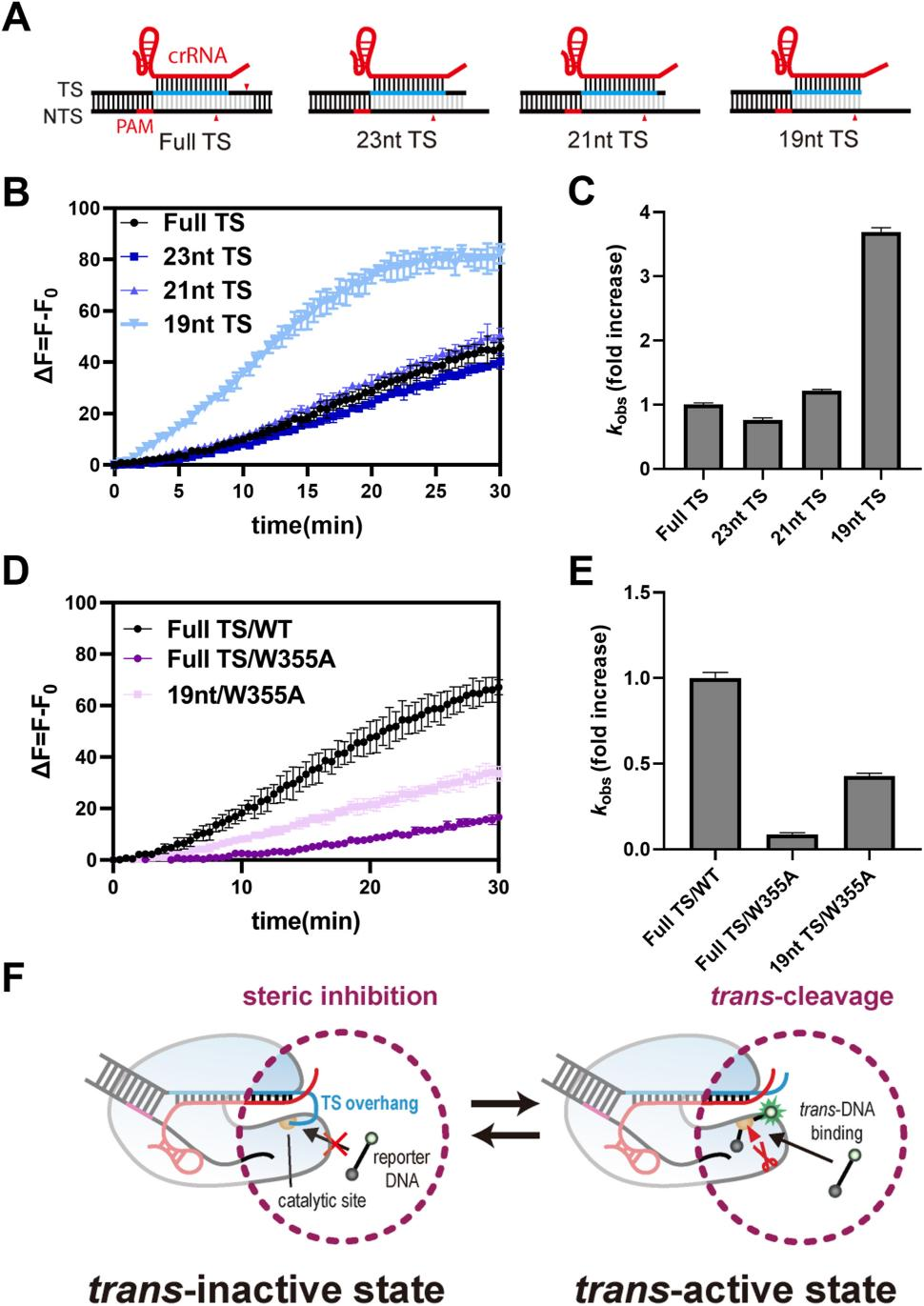
Figure 1: Disruptive Effect of Steric Hindrance from Cis-Cleavage Products on Cas12a Trans-Cleavage Activity
II. Enhancing Trans-Cleavage Activity by Disrupting the Binding Between the TS Protruding End and the Catalytic Site Through Cas12 Mutants
Studies have shown that the target strand loading (TSL) domain of Cas12a is responsible for guiding and stabilizing the target DNA strand at its catalytic site, while retaining the TS during cis-cleavage.
Based on this mechanism, researchers proposed a strategy to increase trans-cleavage activity by mutating the TSL domain of Cas12a to reduce the binding between the TS protruding end and the catalytic site. They introduced alanine mutations in the TSL domain, replacing several surface-exposed hydrophilic residues (including lysine) with hydrophobic alanine residues.
To test the feasibility of this strategy, the researchers performed fluorescent trans-cleavage experiments on various Cas12a mutants and found that these mutations significantly enhanced trans-cleavage activity. In particular, the KE1096AA mutant showed the highest increase in trans-cleavage activity (5.8-fold).
Furthermore, the researchers tested four other Cas12a mutants with hydrophobic residues at positions 1096 and 1097 (KE1096II, KE1096LL, KE1096VV, KE1096FF) and found that their trans-cleavage activities were similar to the KE1096AA mutant.
These results collectively confirm that introducing hydrophobic residues in the TSL domain to create Cas12a variants can significantly enhance Cas12a's trans-cleavage activity while maintaining its cis-cleavage activity.
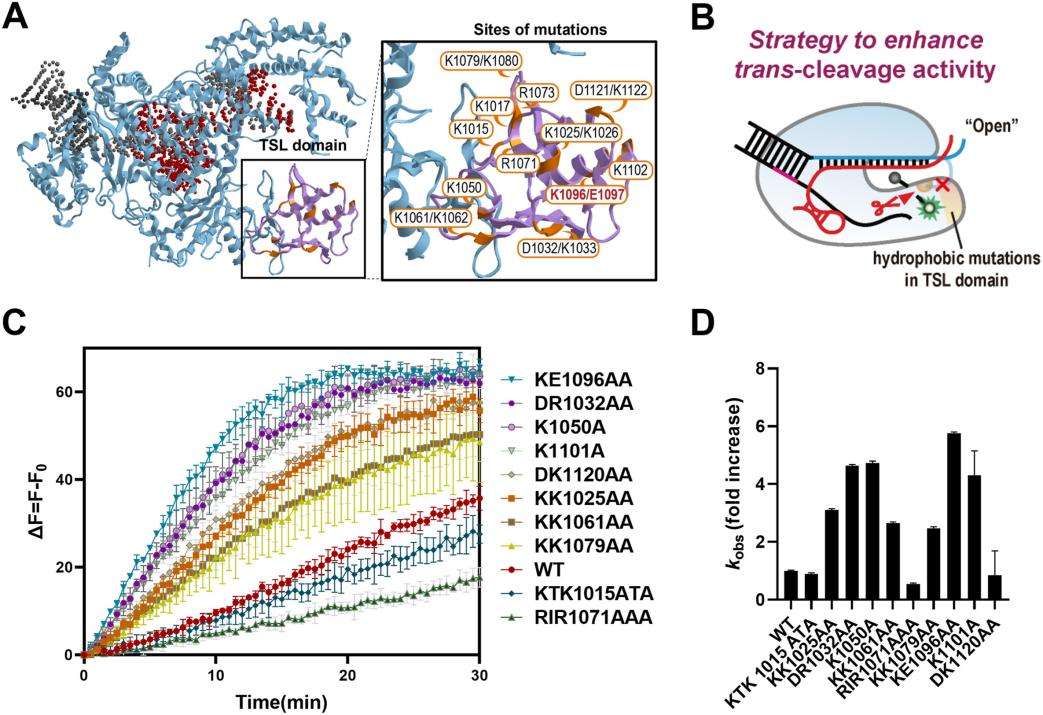
Figure 2: Engineering Cas12a Mutants with Enhanced Trans-Cleavage Activity
III. Enhanced Trans-Cleavage Activity of Cas12a Under Low Ionic Strength Conditions
Due to the polyelectrolyte properties of nucleic acids, the ionic strength of the reaction buffer can significantly impact the in vitro activity of many enzymes, particularly the concentration of monovalent ions (such as Na+ and Cl-), which can greatly affect the molecular interactions between protein domains and single-stranded DNA substrates.
To further enhance the trans-cleavage activity of the improved Cas12a variants, researchers explored optimizing ionic conditions to boost trans-cleavage efficiency. They developed a new strategy called the "salt dilution method," which involves performing cis-cleavage reactions at high salt concentrations and then diluting the salt to enhance trans-cleavage activity while maintaining cis-cleavage activity.
In titration experiments using the salt dilution method, both the KE1096AA mutant and wild-type Cas12a showed increased trans-cleavage activity as NaCl concentration decreased. Under low-salt conditions, the trans-cleavage activity of the KE1096AA mutant was enhanced more than 7-fold.
Additionally, similar effects on trans-cleavage activity were observed with different monovalent ions (including KCl, LiCl, and RbCl). Using this salt dilution method, the researchers successfully enhanced Cas12a activity, significantly increasing the sensitivity of the detection system.
Figure 3: Salt Dilution Method Enhances Cas12a Trans-Cleavage Activity
IV. Detection of Hepatitis B Virus Using Improved Cas12a Mutants and Salt Dilution Method
Using the improved Cas12a mutant (specifically KE1096AA) and the salt dilution method, researchers further evaluated the sensitivity of their developed CRISPR/Cas12a-based system for detecting Hepatitis B Virus (HBV). They assessed the system by testing a series of diluted DNA plasmids containing the HBV polymerase sequence, combined with Loop-Mediated Isothermal Amplification (LAMP) for detection. The results showed that the detection system using the KE1096AA Cas12a mutant and salt dilution method was able to detect the DNA target at the attomolar (atto-M) level, significantly enhancing sensitivity compared to conventional Cas12a-based detection systems.
The researchers also evaluated the sensitivity of the HBV detection system without the DNA amplification step, comparing it to the conventional Cas12a-based method. The results demonstrated a notable increase in sensitivity with the improved Cas12a-based system, even in the absence of DNA amplification.
Additionally, the specificity of the improved Cas12a-based detection system was tested. The researchers used non-specific DNA activators and mixtures containing human genomic DNA to assess specificity, comparing the results with the conventional Cas12a-based method. The findings consistently indicated that the introduced alanine mutations and salt dilution process did not compromise the system’s target specificity.
These results highlight the potential of the proposed Cas12a-based detection system for practical applications, particularly in the rapid and accurate detection of pathogenic nucleic acids.
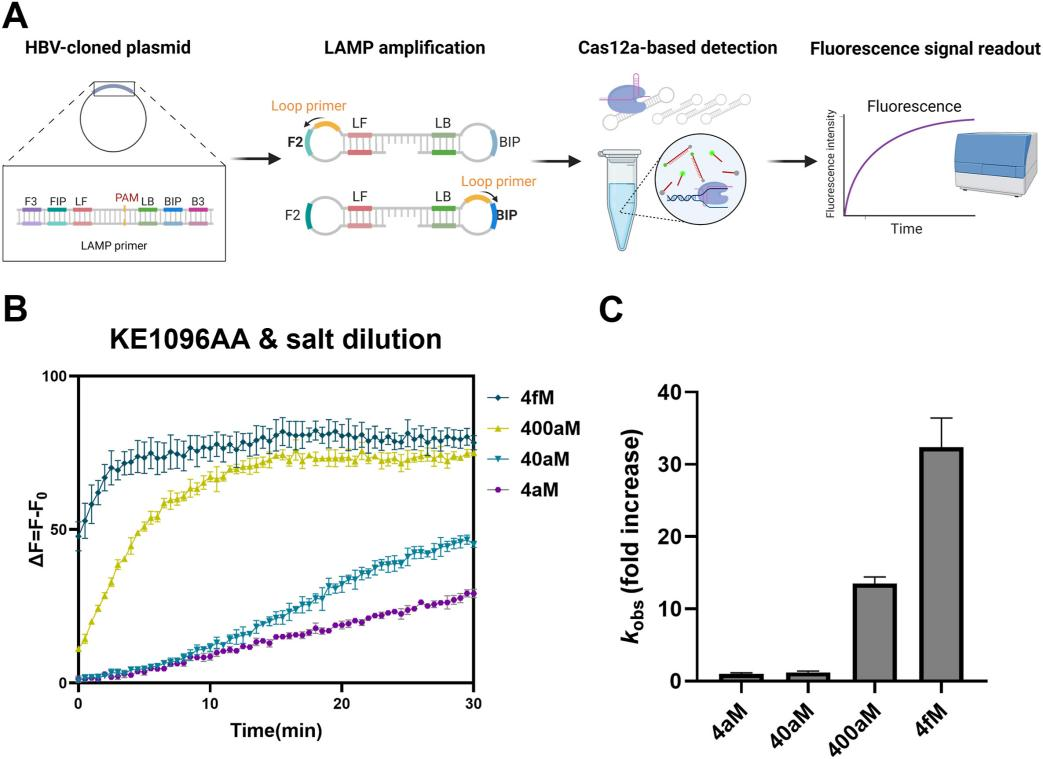
Figure 4: Performance Evaluation of the Improved Cas12a-Based Detection Method
Overall, this study successfully developed a Cas12a variant with enhanced trans-cleavage activity by introducing alanine mutations in the TSL domain of Cas12a. Additionally, a new salt dilution method further boosted Cas12a's trans-cleavage activity under low ionic strength conditions, significantly improving the sensitivity of molecular diagnostics.
The detection method established based on these improvements demonstrated high sensitivity and specificity in diagnosing Hepatitis B Virus (HBV) DNA, showcasing its potential for clinical applications.
EDITGENE boasts its own independently developed protein purification platform.Our Cas enzymes have demonstrated significantly superior activity compared to other market enzymes through rigorous testing, consistently achieving the highest standards in purity, activity, and sensitivity.
Be on the lookout for our isothermal amplification kit and CRISPR detection kits, efficient with minimal equipment requirements and short reaction times—Coming soon!
Recent Blogs
1.[Literature Review] Gene Knockout of ERAP1 Provides New Insights for Optimizing Tumor Immunotherapy
2.[Weekly News] CRISPR Screening Revealing TMEM106B as a Mediator of an ACE2-Independent Infection Pathway for SARS-CoV-2
3.[Weekly News]CRISPR/Cas9 Gene Knockout: A New Strategy to Overcome Immune Therapy Resistance in Neuroblastoma
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com













![[Literature Review] Unlocking CRISPR-Cas12a's Full Potential: Enhancing Trans-Cleavage with Variants](/uploads/20241113/VUPcoYpt8KGLuW0A_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)