[Literature Review] CRISPR Knockout Library Screening Identifies TMEFF1 as a Neuron-Specific Restriction Factor for HSV-1
CRISPR library screening
The CRISPR-Cas9 whole-genome knockout library controls the MOI (Multiplicity of Infection) at 0.3-0.4 to infect cells, ensuring that each cell receives only one sgRNA. This approach allows for the knockout of tens of thousands of genes across different cells simultaneously, playing a crucial role in gene function research, disease mechanism studies, drug development, gene regulation research, and virus-host interactions.
In July 2024, a research team from China, in collaboration with Søren R. Paludan's team from Aarhus University, published significant findings in Nature. By screening the whole-genome knockout library, they identified genes related to HSV-1 virus invasion and replication and explored neuron-specific antiviral mechanisms, opening up new directions for research on combating HSV infections.

Original Article Link:https://doi.org/10.1038/s41586-024-07670-z
Herpes Simplex Virus Type 1 (HSV-1) is a highly prevalent neurotropic double-stranded DNA virus that can cause Herpes Simplex Encephalitis (HSE). HSV-1 infection typically begins in epithelial cells and then spreads to sensory neurons. In most cases, viral replication is controlled, and the virus enters a latent phase. In rare cases, HSV-1 can travel from peripheral nerves into the central nervous system (CNS), leading to HSE.
Cells in the CNS are separated from the rest of the body by the blood-brain barrier, which limits the entry of immune cells and inflammatory components into the CNS. At the same time, neurons are highly sensitive to excessive inflammation and have limited self-renewal capacity. These factors suggest that neurons possess antiviral mechanisms to directly restrict viral replication without activating inflammation. To study the mechanisms by which neurons combat HSV-1, researchers conducted a whole-genome screening, with in vitro and in vivo validations, identifying TMEFF1 as a restriction factor that limits HSV-1 replication.
I. Whole-Genome CRISPR Screening
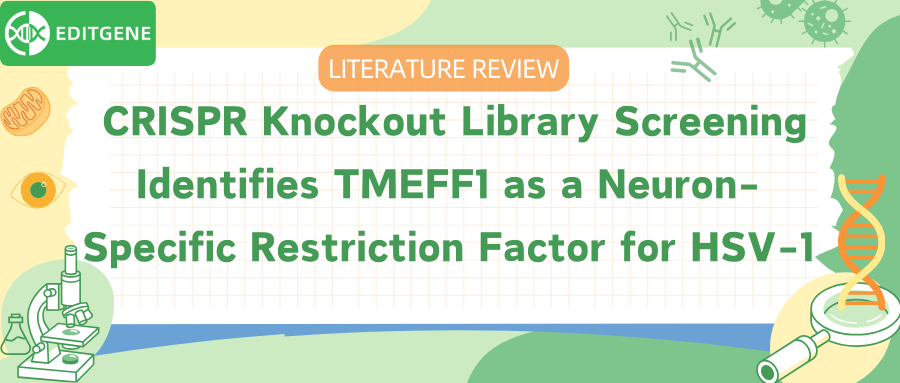
To establish an ideal screening model, the researchers first conducted a preliminary experiment. The results showed that the neuroblastoma cell line SK-N-SH had a limited response through the nucleic acid sensing pathway and did not generate an IFN response to HSV-1 infection. Therefore, this cell line was selected as the screening model. The team then created a stable SK-N-SH Cas9-expressing cell line and infected it with the GeCKO v2 CRISPR library. After 7 days of puromycin selection, they obtained a knockout cell pool and a pool of knockout cells infected with the HSV-1-GFP virus strain. Fluorescent sorting was used to isolate cells with different fluorescence intensities, and MAGeCK was employed to compare the sgRNA enrichment levels between the strongly fluorescent cells and the unsorted cells.
Through this screening process, the researchers identified 322 candidate genes, among which TMEFF1, NKX2-8, and CTXN1 were predominantly expressed in neural tissue. These three candidate genes were then selected for further validation.
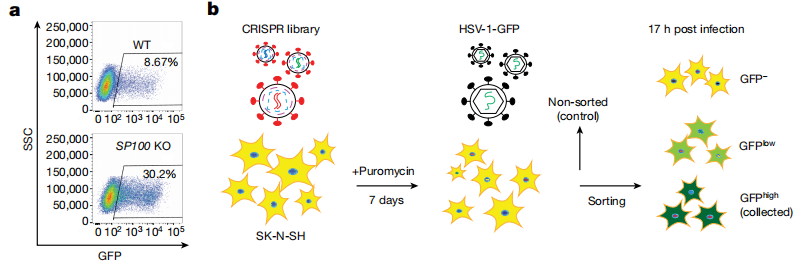
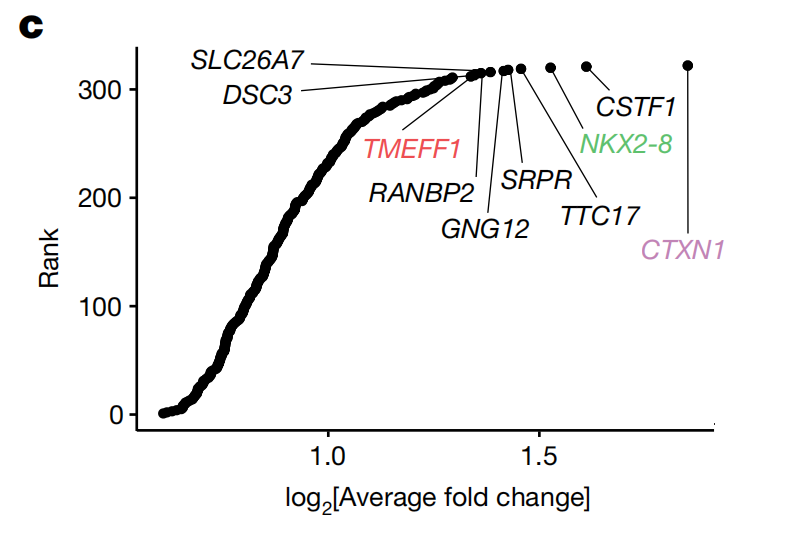
The whole-genome library screening identified genes such as TMEFF1, NKX2-8, and CTXN1
2. Candidate Gene Validation – Tmeff1−/− Cell Model Validation
The researchers further validated the functions of these candidate genes and found that TMEFF1 was the most effective in limiting HSV-1. By knocking out or overexpressing these genes in SK-N-SH cells, they assessed the impact on HSV-1 replication. The results showed that the deletion of TMEFF1 led to a significant increase in HSV-1 replication in neurons and a higher neuronal death rate.
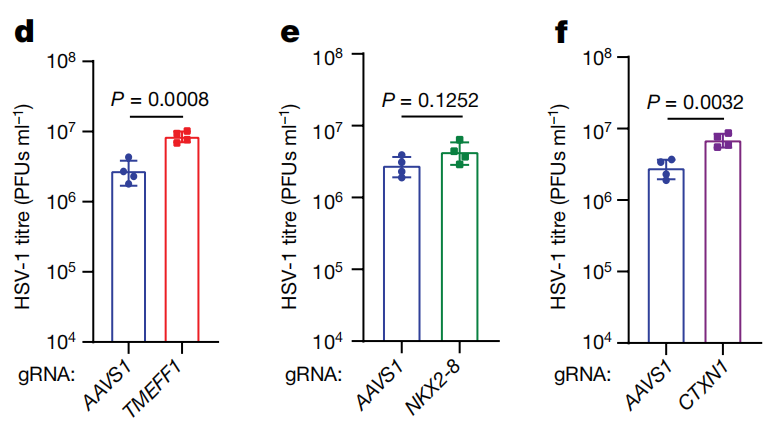
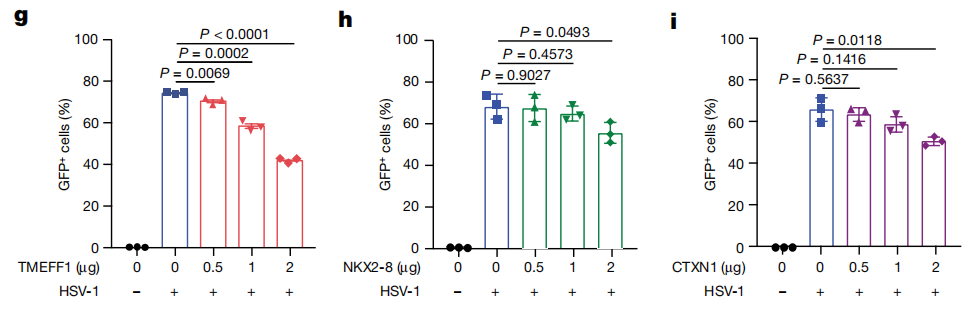
TMEFF1 is a restriction factor for HSV-1 replication in SK-N-SH cells
Using CRISPR-Cas9 technology, TMEFF1 was knocked out in neurons derived from LUHMES embryonic neural progenitor cells and cortical neurons derived from human embryonic stem cells (hESCs), followed by infection with HSV-1-GFP. The results showed that in neurons lacking TMEFF1, the replication efficiency of HSV-1 was significantly increased. The expression levels of the viral genome were notably higher at early stages, accompanied by a higher neuronal death rate. Moreover, these neurons were specifically more sensitive to HSV infection, while there was little effect on Zika virus or poliovirus infections.
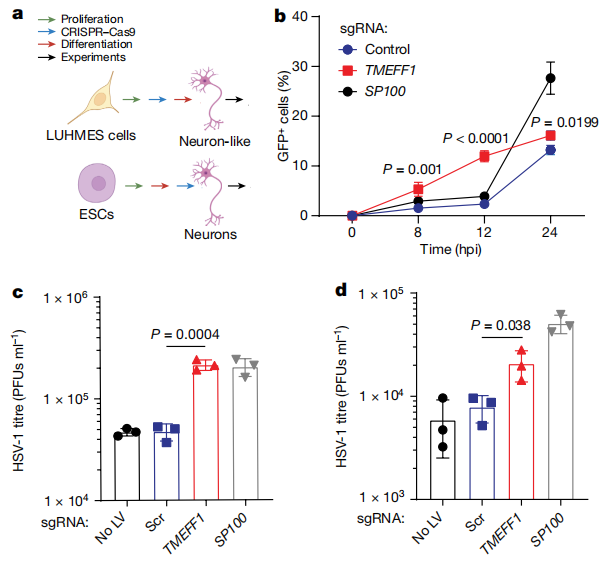
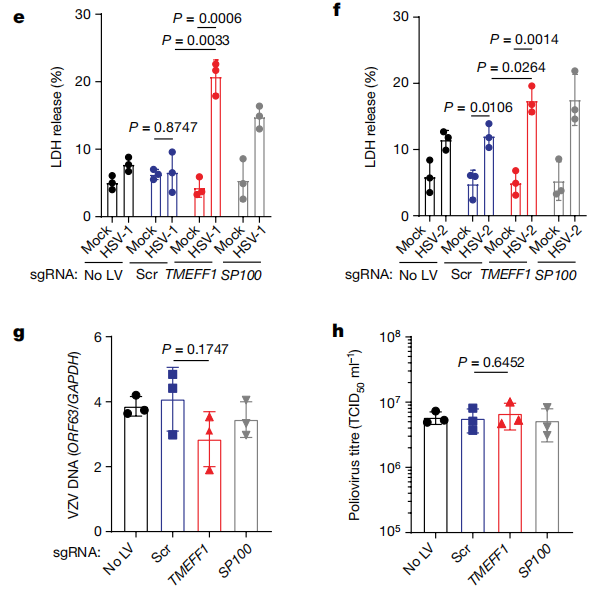
Knockout of the TMEFF1 gene in cortical neurons of nerve cells can affect HSV-1 replication
3. Tmeff1−/− Mouse Model Validation
The researchers used Tmeff1 gene knockout (Tmeff1−/−) mice for HSV-1 infection experiments to observe the mice's survival rates, weight changes, and disease symptoms. After HSV-1 infection, Tmeff1−/− mice showed significantly lower survival rates, faster weight loss, and more severe disease symptoms. Particularly in the brainstem, HSV-1 load was significantly increased, and this increase was mainly observed in neurons.
Immunohistochemical staining revealed that the proportion of HSV-1-positive neurons in the brainstem of TMEFF1-deficient mice was significantly higher, while the proportion of HSV-1-positive astrocytes showed no significant change. This further confirmed that TMEFF1 specifically restricts HSV-1 infection in neurons.
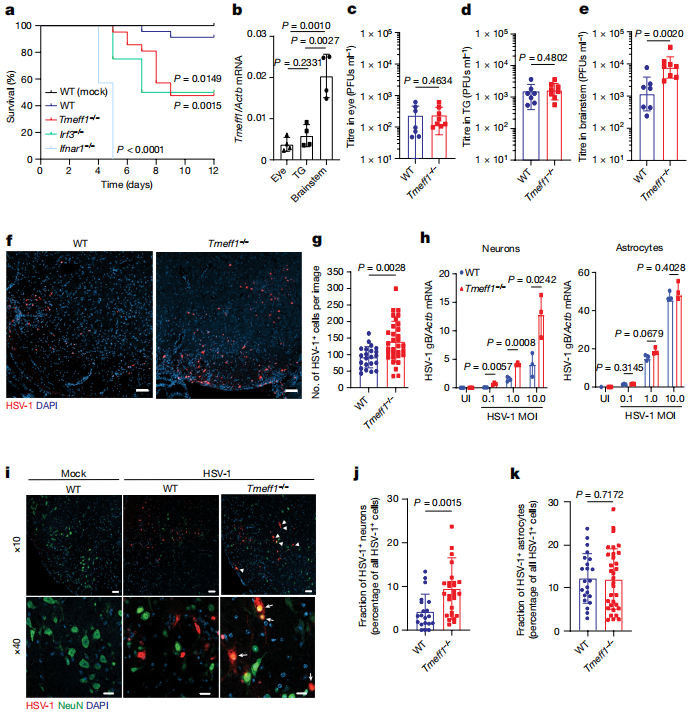
Gene knockout (Tmeff1−/−) mice validate that Tmeff1 is a restriction gene for HSV-1
II. Study on the Antiviral Mechanism of the TMEFF1 Gene Against HSV
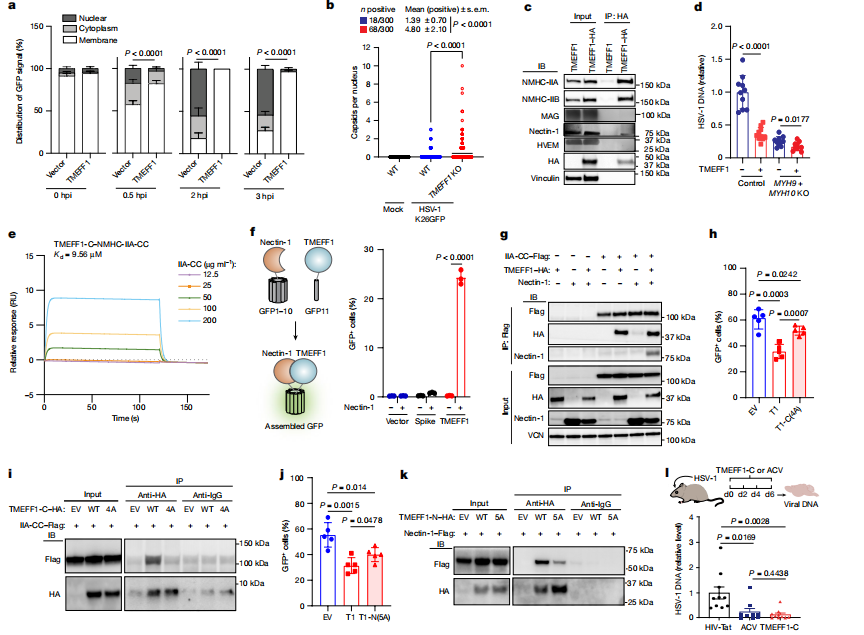
To investigate the viral entry mechanism, TMEFF1 was expressed in HEK293T cells, which were then infected with wild-type HSV-1 and entry-deficient mutant strains (ΔgD and ΔgB). The viral entry into the cells was assessed. In TMEFF1-overexpressing cells, HSV-1 DNA levels in both the cytoplasm and nucleus were significantly reduced, and the accumulation of the viral capsid protein VP5 in the cytoplasm was also decreased.
Shortened and point mutations of TMEFF1 were constructed to examine their interactions with NMHC-IIA and nectin-1, as well as their inhibitory effects on HSV-1 infection. The N-terminal and C-terminal truncated mutants of TMEFF1 showed weakened interactions with nectin-1 and NMHC-IIA, and these mutants exhibited a significant reduction in their ability to inhibit HSV-1 infection.
Co-immunoprecipitation and mass spectrometry techniques were used to identify proteins that interact with TMEFF1. It was found that TMEFF1 interacts with non-muscle myosin heavy chains IIA and IIB (NMHC-IIA and NMHC-IIB) as well as nectin-1. These proteins play key roles in the process of HSV-1 entry into cells.
 TMEFF1 prevents HSV-1 entry by interacting with nectin-1, NMHC-IIA, and NMHC-IIB
TMEFF1 prevents HSV-1 entry by interacting with nectin-1, NMHC-IIA, and NMHC-IIB
Based on this researches, it was found that TMEFF1 prevents HSV-1 entry by interacting with nectin-1 (the gD receptor for HSV-1) and non-muscle myosin heavy chains IIA and IIB (NMHC-IIA and NMHC-IIB, which are involved in the fusion process of HSV-1 entry into cells). Specifically, TMEFF1 acts in the early stages of viral entry by interfering with the fusion of the virus with the cell membrane, preventing the viral genome from entering the cytoplasm and nucleus. TMEFF1 not only limits nectin-1-mediated HSV-1 entry but also inhibits HVEM (another HSV-1 entry receptor)-mediated entry, with a more significant effect on nectin-1-mediated entry. This indicates that TMEFF1 works through multiple mechanisms to effectively restrict HSV-1 infection.
In onclusion, this study systematically demonstrated the key role of TMEFF1 in restricting HSV-1 infection in neurons through in vitro and in vivo experiments. It revealed that TMEFF1 blocks HSV-1 entry into cells by interacting with nectin-1 and NMHC-IIA/IIB. These findings not only provide new insights into the mechanism of HSV-1 infection in the nervous system but also offer potential avenues for developing new therapies against HSV-1.
EDITGENE offers a one-stop comprehensive solution for CRISPR library screening, including sgRNA library design and customization, Cas9 stable cell line construction, library lentivirus packaging, library cell generation, functional screening experiments, and NGS analysis. We provide popular libraries such as GeCKO v2 CRISPR and offer the most complete collection of Library Plasmids and library viruses in stock, with delivery within one week. Order now and start screening immediately!
Recent Blogs
- 1. [Weekly News] Breakthrough in Gene Editing: Overcoming Immunogenicity with Cas Enzyme Mutants
- 2. [Weekly News] CRISPR Library Screening Uncovers ALCAM as a Novel Molecular Target for Human Adenovirus B
- 3. [Weekly News] CRISPR-StAR: High-Resolution Genetic Screening Innovation
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com












![[Literature Review] CRISPR Knockout Library Screening Identifies TMEFF1 as a Neuron-Specific Restriction Factor for HSV-1](/uploads/20250208/Rpauo17ZWDsPwzIl_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)