Prime editing 101:what is prime editing?
Prime editing is an advanced gene editing technology proposed by David R. Liu's team in 2019. It can mediate targeted insertions, deletions, and twelve base substitutions without DSBs and donor DNA templates to achieve precise gene editing. Prime editing has developed rapidly in the past 4 years and has shown great potential in various fields.
Prime Editing mechanism
The prime editing system consists of Prime editing guide RNA (pegRNA) and nCas9. PegRNA can be divided into a guide RNA at the 5' end, a Cas9 binding region in the middle, and a PBS (Prime-binding site) at the 3' end; nCas9 is a Cas9 nickase with single-stranded cleavage activity and fused with M-MLV reverse transcriptase.
The complex recognizes PAM under the g uidance of pegRNA and binds to the target DNA strand complementary to the guide RNA. Cas9 enzyme cuts the 3rd and 4th nucleotides of the original spacer sequence upstream of PAM to generate a nick; the broken DNA single strand binds to PBS. , the reverse transcriptase takes effect and performs reverse transcription along the PBS to generate the editing sequence; the editing sequence and the original sequence of the target DNA will compete with the other strand through base pairing, and the unbound sequence will become tilted and finally cut off.
Competitive binding will eventually produce two results: one is that the original sequence of the target DNA is left and the editing sequence is removed. At this time, the target DNA is not successfully edited and may be recognized by the complex again; the other is that the editing sequence is left. Next, the original sequence of the target DNA is removed. At this time, because the edited sequence does not match the sequence of the other strand, a protrusion is formed at the editing site, and the DNA mismatch repair mechanism in the cell repairs it. The repairing results are also divided into two types, one is that the editing site is modified and returns to the original state of the target DNA; and the other strand is repaired and the edited DNA double strand is produced, successfully achieving the purpose of precise gene editing.
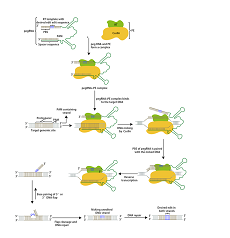
Figure 1 Point Mutation
The first batch of Prime Editing data released
On October 27, 2023, Dr. Jeremy S Duffield, Chief Scientific Director of Prime Medicine, reported that Prime editing precisely corrects prevalent pathogenic mutations causing Glycogen Storage Disease Type 1b (GSD1b) at the European Society of Gene and Cell Therapy in Brussels.
Glycogen storage disease (GSD) is a genetic metabolic disease caused by defects in glycogen decomposition or synthase enzymes. The SLC37A4 gene mutation of glucose-6-phosphate translocase (G6PT) leads to GSD type Ib. In the fasting state, The inability to produce glucose causes hypoglycemic episodes and related seizures and causes immune disorders characterized by neutropenia and myeloid dysfunction. In the population, 50% of GSD1b patients have the SLC37A4 gene mutation p.L348fs — and p.G339C. The latest research and development data from Prime Medicine shows that its Prime Editors accurately corrected the type 1b glycogen storage disease p.L348fs mutation in humanized mice, with an accurate correction rate of more than 56% in the whole liver, and precise editing of the G6PT gene restored G6PT Protein expression; Efficient and accurate editing of the SLC37A4 (G6PT) gene p.L348 in the liver of large animal cynomolgus monkeys, with up to 50% accurate editing of the whole liver and good tolerance by cynomolgus monkeys.

Figure 2 Introduction of the speech
The development of Prime Editing
The original Prime editing system was called PE1, and the editing efficiency was not very high, and there may be non-specific. Later, to improve the editing efficiency, a series of optimized systems were developed.
PE2: Introduce several mutations into M-MLV reverse transcriptase, so that the reverse transcriptase has better activity under high-temperature conditions and binds more tightly to pegRNA. The final editing efficiency is 1 to 5 times higher than PE1.
PE3: Add new sgRNA to guide Cas9 to cut DNA. Ideally, it can recognize DNA that has been edited but has not been repaired by a mismatching mechanism and guides the Cas enzyme to cut on the DNA chain complementary to the editing site to improve editing efficiency. But in fact, nicks are often produced on both DNA strands, inducing the mismatching mechanism and introducing indels.
PE3b: The editing sequence will be added to the newly designed sgRNA to achieve ideal conditions, repairing the DNA strand complementary to the editing site.
PE4, PE5: Studies have found that the PE editing efficiency is reduced when DNA is repaired for mismatches, and it will stimulate the production of indels. PE4 (PE5) is based on PE2 (PE3) and transiently expresses a dominant loss-of-function (dn) protein MLH1dn. The protein lacks three amino acids at the very end of the carboxyl terminus, causing it to lose its endonuclease function. , transiently inhibiting the activity of mismatch repair enzymes.
PEmax: There are a series of improvements and optimizations based on PE2, such as optimizing reverse transcriptase codons, mutating some amino acids in SpCas9 to improve enzyme cutting efficiency, and adding NLS peptides to both ends of the entire enzyme to increase the adsorption of nucleic acids by the complex enzyme. These optimizations increased the editing efficiency of PEmax by 2.5 times in Hela cells and 1.2 times in HEK293T cells compared with PE2.
EDITGENE optimized and upgraded Bingo ™ prime editing point mutation platform , using the latest PEmax system to provide precise and efficient gene point mutation cell constructionservices for the many scientific research enterprises.
Prime Editing Applications
Currently, Prime editing has been widely used in research on disease treatments.
In the phenylketonuria (PKU) Pahenu2 mouse model, human adenoviral vector 5 (AdV) was used to deliver PE3 Δ RnH to the Pahenu2 mouse model, with an average correction efficiency of 11.1% and no off-target mutations or other liver inflammation detected, indicating The prime editing system plays a role in the clinical treatment of PKU;
The human Leber congenital amaurosis (LCA) mouse model was injected with split AAVE-PE2 to repair the nonsense mutation (C → T) in RPE65 exon 3, with an average editing efficiency of 28% and no unintended edits;
The Prime editing system was used to insert 2 bases into exon 52 of the Δ Ex51 iPSC model to treat Duchenne muscular dystrophy (DMD), with an editing efficiency as high as 54%.
Providing a ccurate treat ment to genetic diseases caused by indels, Prime Editing is feasible to customize gene therapy with its precise and efficient gene repair characteristics. With the continuous improvement and improvement of technology, Prime Editing is expected to become an important tool for future gene therapy and precision medicine, bringing new hope to human health.
Bingo ™ P rime Editing Point Mutation Platform
Based on PE (Prime Editing) technology, EDITGENE has newly upgraded and developed the Bingo™ prime editing point mutation platform. W e summarized, optimized and improved the experience of thousands of gene editing CRO projects. The success rate far exceeds the traditional gene point mutation method.
Contact Us
Email:info@editxor.com
Phone:833-2263234 (USA)
Recent Posts:
[Research Frontier] New trends in CRISPR detection: Monkeypox virus detection
【Monthly Star】AMPK,TREM2 and p53 KO cell lines
[Research frontier] CRISPR Detection Trends










![[Literature Review] A Novel Mechanism of Cisplatin Resistance in Osteosarcoma: CRISPR Screening Identifies Key Regulators in Organoid Models](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)
![[Quality Share] Decoding Point Mutations: A Comprehensive Guide to Three Common Construction Methods](/uploads/20250328/ESzk5OC49wpxIHVv_3cbfa5e98ea1d238127fe23c72b0f4b2.png)

Comment (4)