[Literature Review] Improving the Sensitivity of LbuCas13a Detection using Engineered crRNAs
The CRISPR system is part of an adaptive immunity system in bacteria and archaea. Cas nuclease guided by crRNA can be edited to recognize the desired target sequence for further gene editing or nucleic acid detection. There are several methods to improve the detection ability of CRISPR systems, such as fusing RNA binding domains to the Cas13a active site, designing hairpin reporters, integrating multiple crRNAs from different nucleotide regions of a single nucleic acid, or crRNA engineering modifications. crRNA engineering is the simplest and most cost-effective method. It has been used to improve the Cas9 and Cas12a nuclease's cis-cleavage activity, thereby improving the efficiency and specificity of gene editing. Still, the use of engineered crRNAs to enhance nucleic acid detection in the Cas13a system has not been reported to date.
In March, researchers from the State Key Joint Laboratory of ESPC at Tsinghua University and the Beijing Center for Disease Control and Prevention published a research paper in Biosensors and Bioelectronics, titled Improving trans-cleavage activity of CRISPR-Cas13a using engineered crRNA with a uridinylate-rich 5′-overhang , the researchers engineered crRNA for Cas13a through various extensions and chemical modifications. Through repeated experiments, they’ve identified the best-engineered crRNA with optimized performance and conditions. They used the modified LbuCas13a system in combination with RT-RPA, or isothermal reverse transcription- recombinase polymerase amplification technology, to detect SARS-CoV-2 RNA within clinical samples, providing a valuable reference for the subsequent development of more efficient Cas13a/crRNA systems.
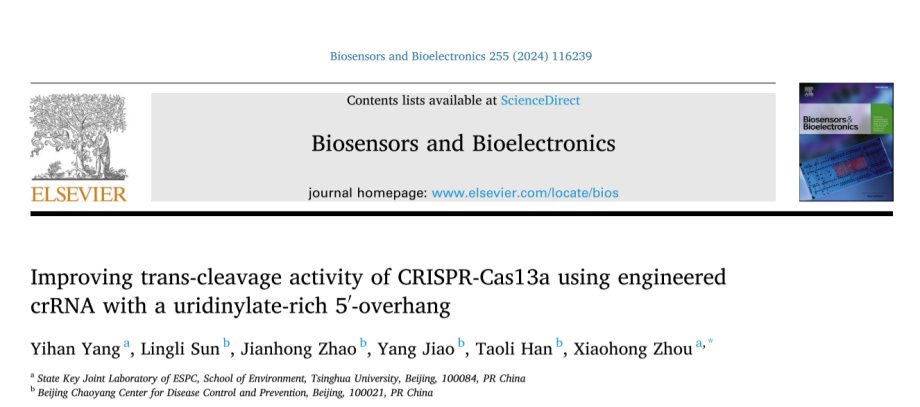
I. Exploring the U extension effects of different lengths and chemical modifications on crRNA performance
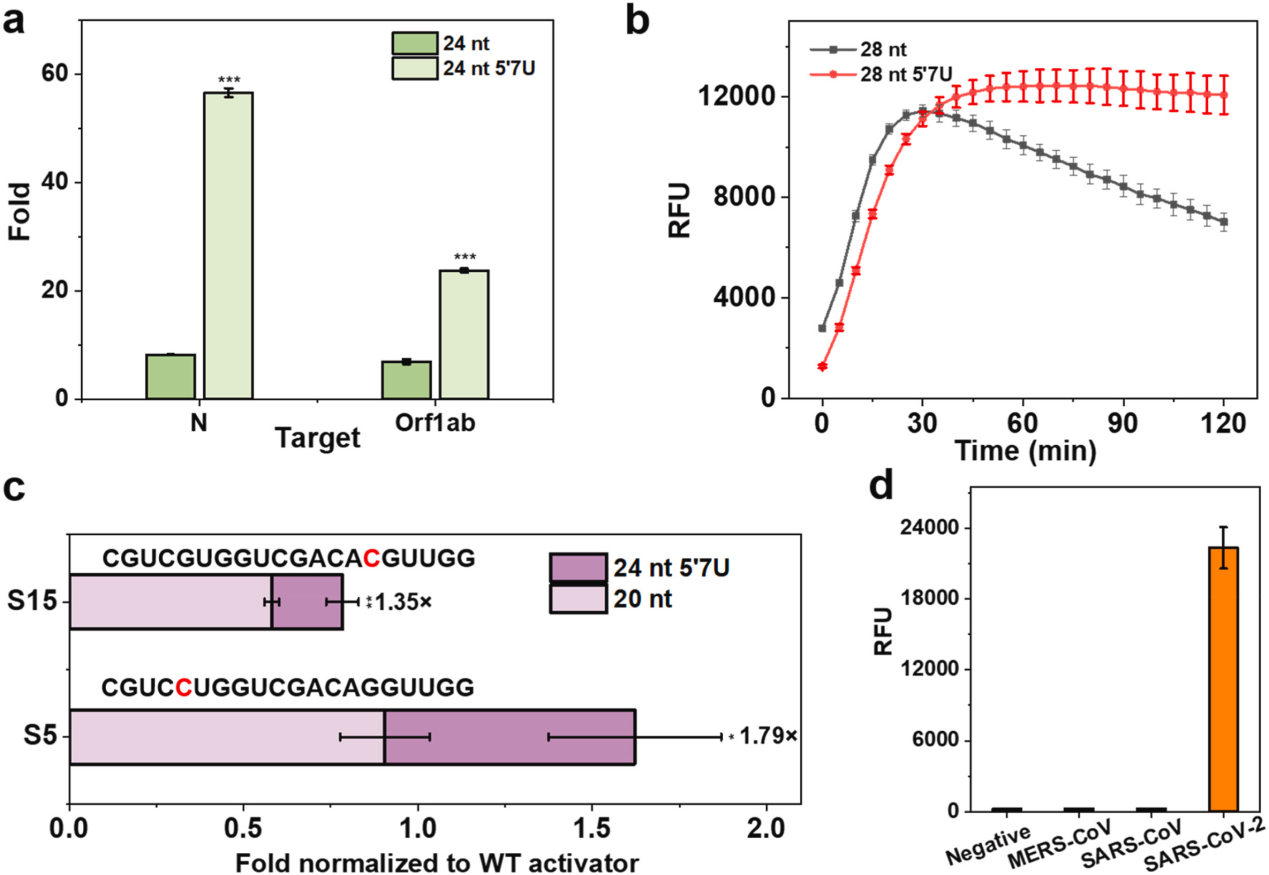
The researchers selected a commonly used crRNA to be engineered by adding different amounts of uracil (U) at the 3′ or 5′ end of the crRNA or through chemical modification to enhance the trans-cleavage activity of Cas13a. The experimental results showed that crRNA with 3U extension at the 5′ end significantly enhanced the trans-cleavage activity of LbuCas13a , whereas the extension of 3U at the 3′ end decreased the activity. Chemical modification of crRNA with PS-2′-OMe at both ends of 3U extension, respectively, resulted in the increased stability of Cas13a but decreased the trans-cleavage activity. Not only so, the researchers also used it to replace the crRNA's 2 ′-OH of cytosine in the repeat or guided region and tested its trans-cleavage activity. They found that crRNAs with cEt substitutions in the repeat region had minimal Cas13a activity and that crRNAs with cEt substitutions in the A3 and G4 guide regions were more active than the original crRNAs. The researchers also explored the effect of different spacer lengths of crRNAs on Cas13a, in which the 24 nt crRNA showed higher activity than the 20 nt crRNA .
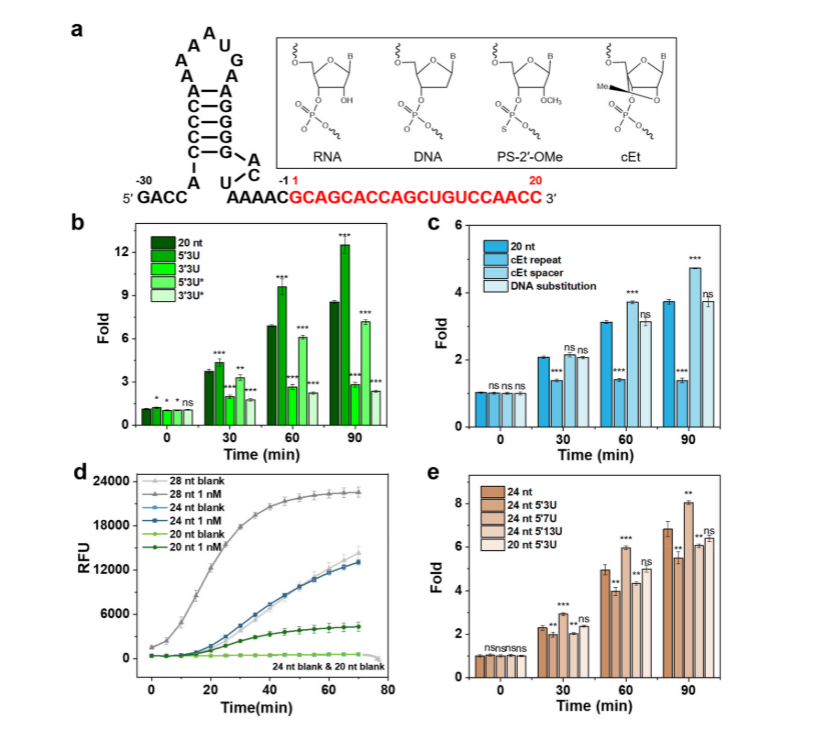
II. Optimizing the binding mechanism of crRNA and LbuCas13a
To investigate the binding mechanism of crRNA and Cas13a, the researchers performed all-atom molecular dynamics simulations (MD) with 20 nt 5′3U, 24 nt, 24 nt 5′7U, and 28 nt crRNA to simulate the trans-cleavage activity of the LbuCas13a protein in the absence/presence of a target. Root-mean-square deviation (RMSD) was used to measure the conformational changes of the MD-simulated system, and it was found that the interactions between the 5′-terminal protrusions and the Helical-1 and HEPN2 structural domains of Cas13a played a key role in the stability of the complexes. The simulation results revealed the effects of crRNA modifications on Cas13a binding and trans-cleavage activity and illustrated why the 5′-terminal extended crRNA can enhance the system's activity. In addition, the researchers performed Michaelis-Menten kinetic studies on 20 nt and 24 nt 5′ 7U crRNAs, which catalyzed trans-cleavage of 20 nt and 24 nt 5′7U crRNAs at a rate of 6.0 and 12.4 rpm, respectively. When the LbuCas13a-crRNAs were bound to the target RNA activator molecules, the calculated catalytic efficiency of 24 nt 5′7U crRNA for LbuCas13a was 3.6-fold higher than that of 20 nt crRNA, suggesting that 24 nt 5′7U crRNA may help to stabilize the Cas13a complex in the optimal conformation for RNA trans-cleavage activity.
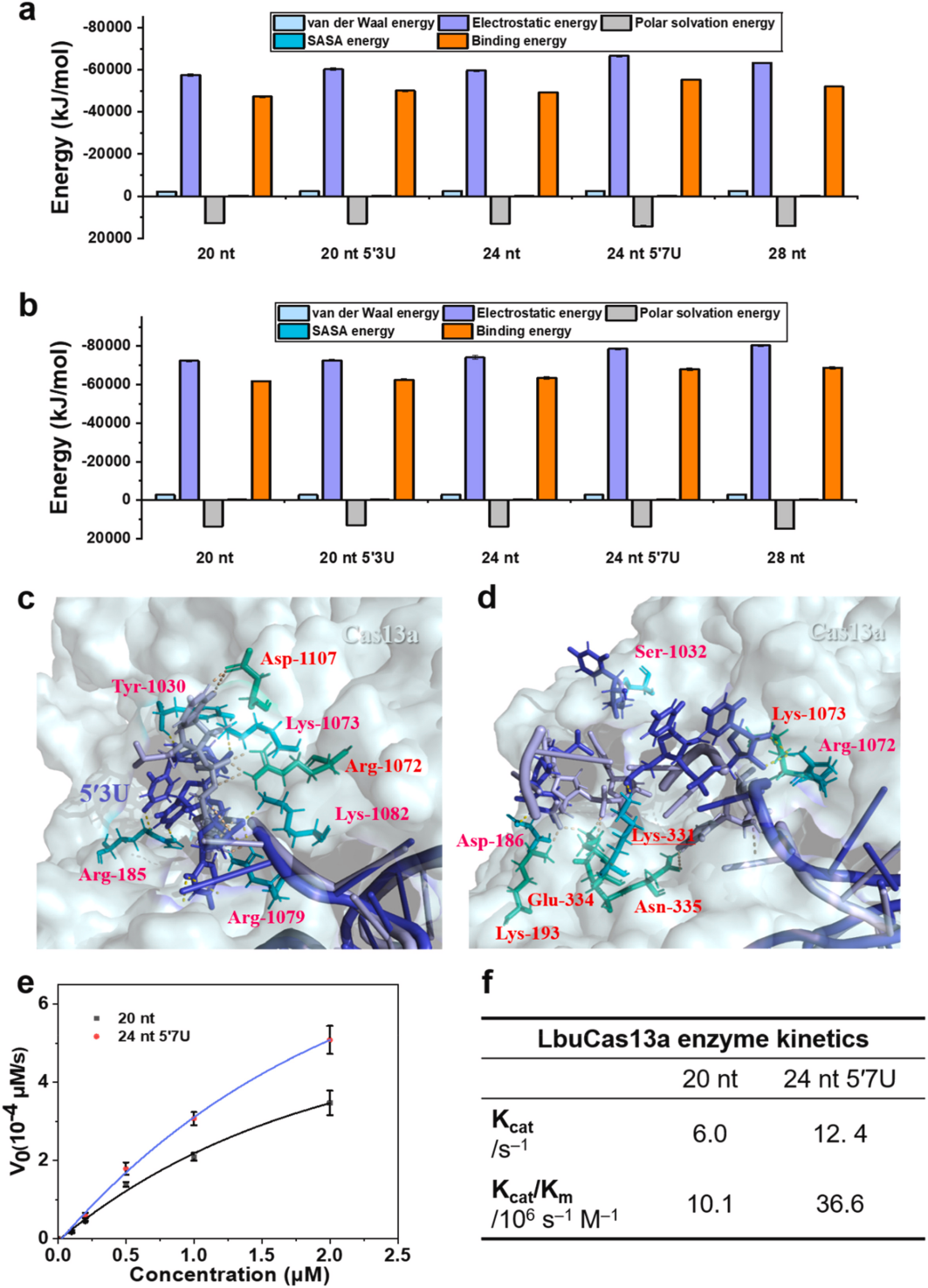
III. Determining the Ionic Dependence of LbuCas13a Trans-Cleavage
To determine the ion dependence of Cas13a trans-cleavage, the researchers tested various divalent ions in side-branch cleavage experiments and found that the trans-cleavage activity of LbuCas13a was enhanced in the presence of Mg2+ , Ca2+ , and Mn2+ , whereas Co2+ , Zn2+ , and Cu2+ significantly inhibited LbuCas13a activity. To investigate the dose-dependence of the inhibitory effects, the researchers determined the effects of Co2+ and Mg2+ concentrations on LbuCas13a trans-cleavage activity, and the results showed that increasing the Co2+ concentration significantly reduced the fluorescence signals of both 20 nt and 24 nt 5′ 7U crRNAs, and that the activity of the 24 nt 5′7U crRNA was higher than that of the 20 nt crRNA at all concentrations. LbuCas13a activity was first enhanced, followed by decreasing effects when Mg2+ concentration was increased. By varying the amount of Mg2+ , the optimal Mg2+ concentration was determined to be 1.5 mM . Heparin was used to avoid nonspecific interactions between the LbuCas13a-crRNA binary complex and ssRNA.Thus, the researchers compared the trans-cleavage activity of the CRISPR-Cas13a system in nuclease assay buffer with and without heparin to validate whether heparin-induced non-specific interactions between crRNA and LbuCas13a-crRNA binary complexes increase the number of LbuCas13a-crRNA complexes and thus enhances the trans-cleavage activity. Polyacrylamide gel electrophoresis showed that crRNA residues were higher in the presence of heparin, suggesting that heparin prevents the interaction of crRNA with the ssRNA binding domains.
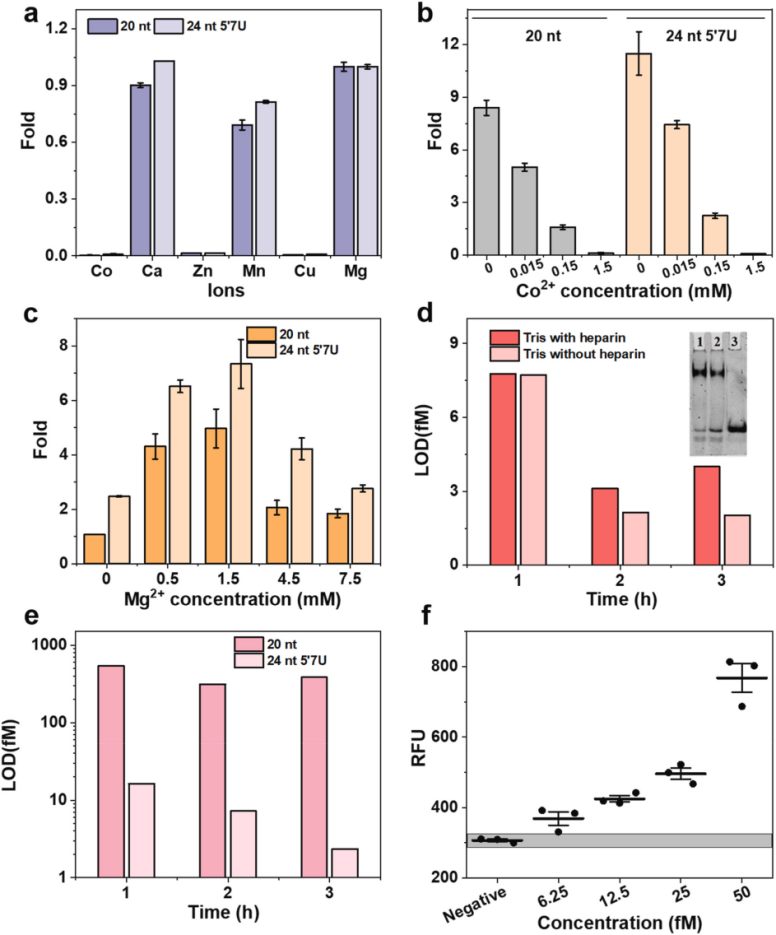
IV Tests to optimize the suitability of crRNAs with CRISPR-Cas13a
The researchers used the same engineering strategy described above to modify crRNA with 7U extension at the 5′ end of the N and Orf1ab genes in SARS-CoV-2. The two engineered systems were 6.88-fold and 3.44-fold more sensitive to the N and Orf1ab genes, respectively, than the unmodified systems and the same experiments were performed by replacing the LwaCas13a, giving similar results. This suggests that the 5′-end extension also favors side branch cleavage in the LwaCas13a system. To further test the specificity of the enhanced CRISPR-LbuCas13a system in recognizing point mutations within the target RNA, the researchers generated two targets, each with a single point mutation in either the seed region (S15) or the spacer region (S5), and were compared to the 20 nt crRNA. The 24 nt 5′7U crRNA was more tolerant to single-nucleotide mismatches in both cases and produced a stronger fluorescence signal.
V. Detection of SARS-CoV-2 Clinical Samples Using an Engineered crRNA Combined with the LbuCas13a System
Researchers established the RT-RPA-amplified 5′7U crRNA CRISPR-LbuCas13a system to detect the S gene of SARS-CoV-2. Rapid detection of the S gene in RNA extracts from 45 clinical samples was performed by a 30-minute RT-RPA reaction at 42°C and a 10-minute CRISPR-Cas13a cleavage reaction. The experiment included one RT-qPCR-validated wild-type SARS-CoV-2-positive sample, 35 RT-qPCR genome-sequencing-validated Omicron-positive variant samples, and 9 RT-qPCR-validated negative samples. Positive and negative controls were tested with synthetic RNA and nuclease-free water. Despite the RNA having been stored for more than two years, the researchers successfully identified wild-type SARS-CoV-2, and the technique accurately identified the Omicron variant of the S gene in the patient samples within 40 minutes with a concordance of 20/21 at a Ct value of < 34 , suggesting that the technique has promising potential for COVID-19 diagnosis.
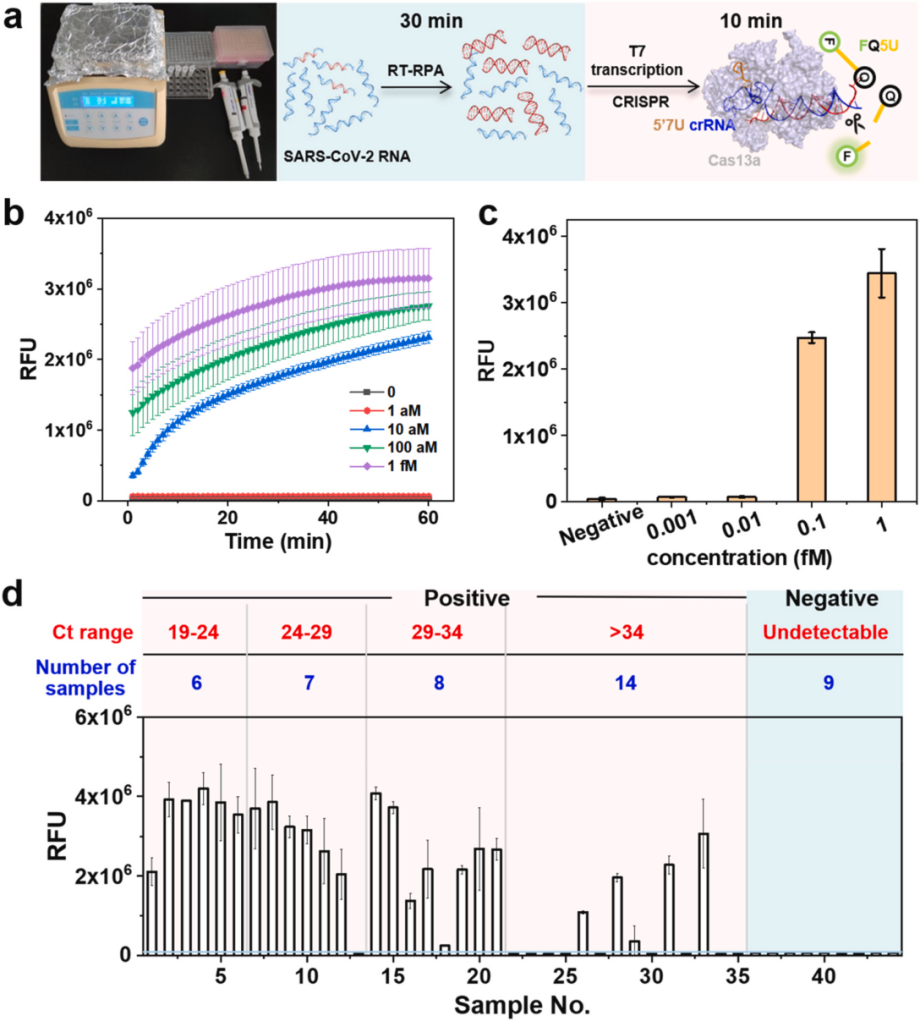
In summary, by engineering crRNA, researchers have significantly improved the sensitivity of the CRISPR-Cas13a system, making it more suitable for detecting samples with low viral loads and combining it with RT-RPA technology to complete the detection of SARS-CoV-2 in a shorter time. This improves the performance of the CRISPR-Cas13a system and provides valuable experience and methods for future engineering and enhancement of other CRISPR systems, making CRISPR-Cas13a system expected to become a powerful tool for POCT diagnosis.
L ink to study : https://doi.org/10.1016/j.bios.2024.116239
EDITGENE has independently developed a protein expression purification platform that has obtained high purity, high activity, and high recovery Cas12/Cas13 enzymes with detection limits up to the amol level! It can be used for CRISPR R&D, genetic testing, animal disease testing, personnel disease testing, and food safety testing.
You might be interested in :
The Knits and Grits behind single-cell CRISPR screening
[MAR Bestselling] CRBN and Myc knockout cell lines; CRISPR whole genome library
[Research Frontier] New trends in CRISPR-- Kras, NLRP3 and BRAF knockout cell lines
Follow us on social media
Contact us:
+1(224) 345-1927 (USA)
+86-13250544625 (Intl)










![[Literature Review] A “Gene Scalpel” Emerges: A New Hope for Tay-Sachs Disease Cure](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)