New Developments in Prime Editing: Phage-Assisted Evolution and Protein Engineering Yield More Efficient and Compact Prime Editors
Prime Editing

Prime Editing, also known as prime editors, is a groundbreaking technology in the field of gene editing that provides a novel tool for precise genetic modifications. Compared to traditional CRISPR-Cas9 technology, PE offers significant advantages in terms of editing accuracy and safety, holding promise for applications in gene therapy, agricultural breeding, and fundamental research.
In August 2023, the journal Cell published an online article titled “Phage-assisted evolution and protein engineering yield compact, efficient prime editors” Conducted by David R. Liu’s team, this research leveraged phage-assisted evolution and protein engineering to develop more compact and efficient prime editors (PE6), marking a new breakthrough in the advancement of gene editing technologies.

Original Article Link:DOI: 10.1016/j.cell.2023.07.039
Highlights:
- a. Phage-Assisted Evolution (PE-PACE): Developed an efficient directed evolution system by directly linking Prime Editing (PE) activity to phage propagation, resulting in smaller and more robust reverse transcriptase (RT) variants.
- b. Ultra-Compact Prime Editor (PE6): Achieved a reduction in RT size to 1.2–1.5 kb through protein engineering and evolution, with a 22-fold increase in editing efficiency, significantly outperforming the traditional M-MLV RT.
- c. Advances in Complex Editing Scenarios: PE6 excelled in tasks such as inserting long fragments and editing templates with high secondary structure, making it particularly well-suited for twinPE editing mediated by dual pegRNAs.
- d. Successful In Vivo Delivery: Using a dual AAV system, PE6 efficiently inserted a 42-bp loxP sequence into the mouse cerebral cortex at a high efficiency (62%), a 24-fold improvement over previous generations.
Prime editing is a DNA double-strand break-free gene editing technology that enables single-base substitutions, small insertions/deletions, and long fragment knock-ins. However, traditional PE, which relies on M-MLV reverse transcriptase (RT), has two major limitations:
(1) Large size: The M-MLV RT gene is approximately 2.2 kb long, making it difficult to fit into AAV and other delivery vectors.
(2) Efficiency bottleneck: Different RTs vary significantly in efficiency for complex edits (such as long fragments or high-structure templates), which constrains clinical applications. To address these issues, the research team combined phage-assisted evolution with protein engineering to develop a next-generation prime editor.
I. Phage-Assisted Evolution (PE-PACE)
The researchers designed a directed evolution system based on T7 RNAP that directly links RT activity to phage propagation (Fig. 1). By applying continuous evolutionary pressure, they identified several highly efficient RT variants:
a. Ultra-compact RT: The Ec48 RT (1.2 kb) achieved a 22-fold increase in efficiency, performing comparably to full-length M-MLV RT.
b. Highly adaptable variants: The PE6d, tailored for long fragments or high-structure templates, surpassed traditional PE3 in complex editing scenarios.
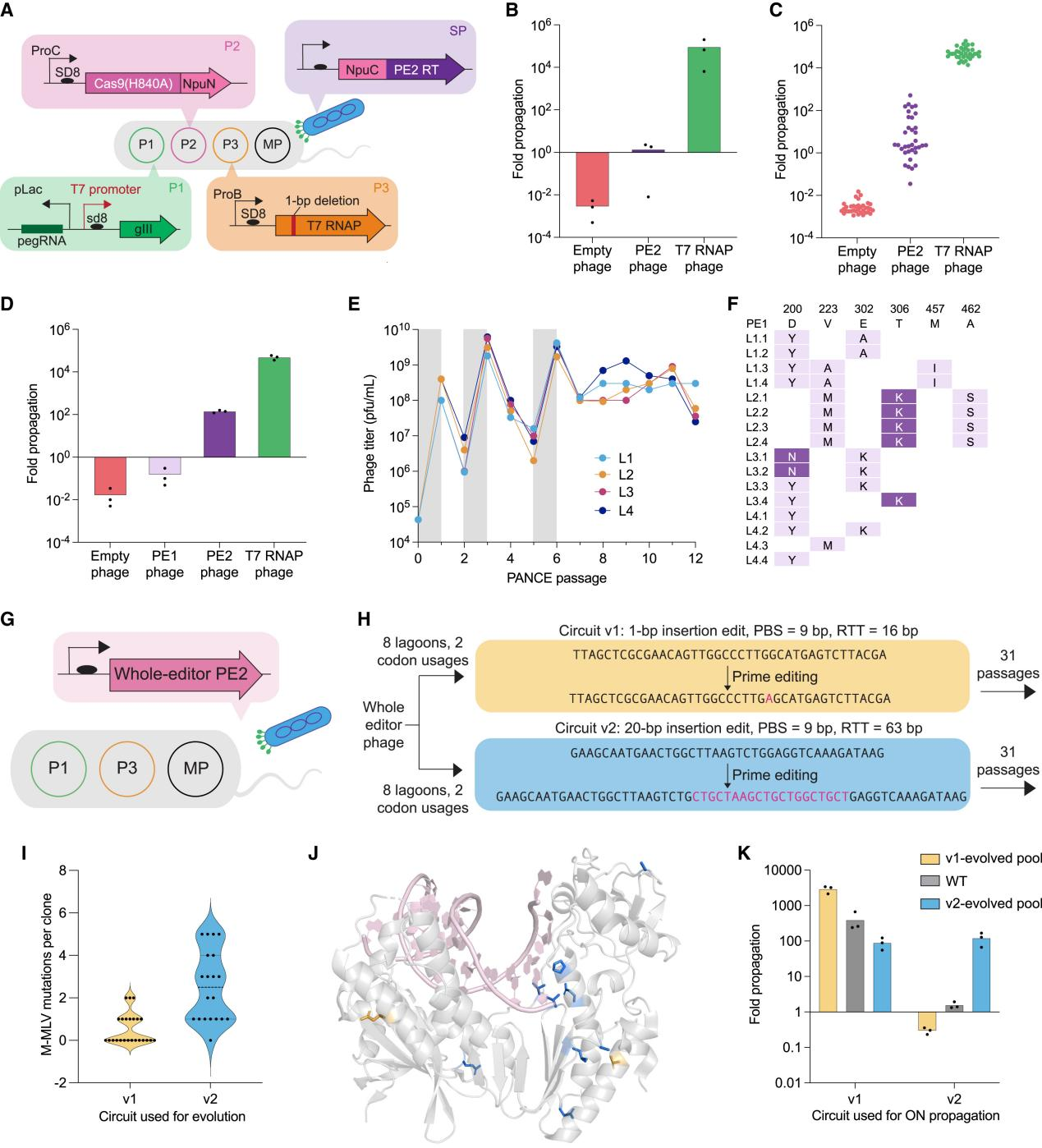 Figure 1: Development and Validation of PE-PACE
Figure 1: Development and Validation of PE-PACE
II. Protein Engineering to Optimize RT
Using AlphaFold-predicted RT structures, the research team introduced mutations in critical substrate-binding domains (Fig. 2), significantly enhancing DNA/RNA binding affinity and processivity. This allowed the compact RT to efficiently handle long-fragment insertions.
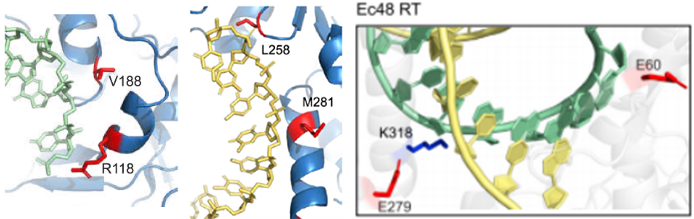
Figure 2: Mutations in the Critical Substrate-Binding Domain of RT
III. Dual AAV Delivery System
By adapting the PE6 editor to a dual AAV system, researchers inserted a 38 bp attB sequence into the mouse cerebral cortex, achieving an editing efficiency of 10.4% in transduced cells (Fig. 3B). They also successfully inserted a 42 bp loxP sequence, with a final efficiency of up to 62% and an off-target rate below the detection limit (Fig. 3C).
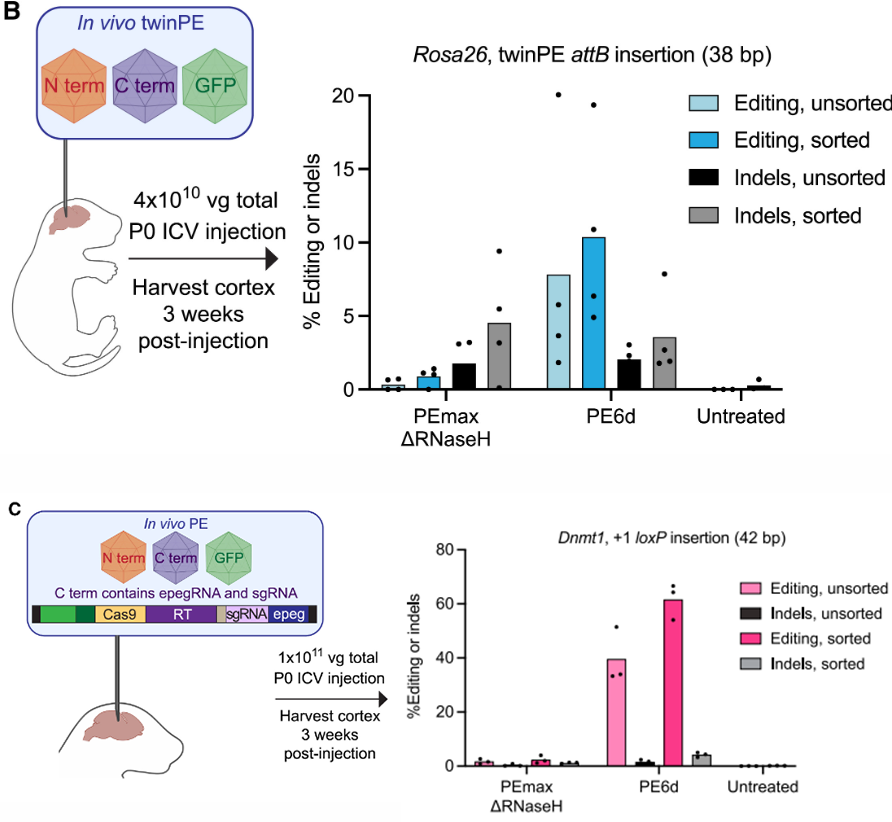
Figure 3: In Vivo Editing Using PE6 Editors
IV. Application Scenarios
1. Point Mutations & Short Fragment Edits
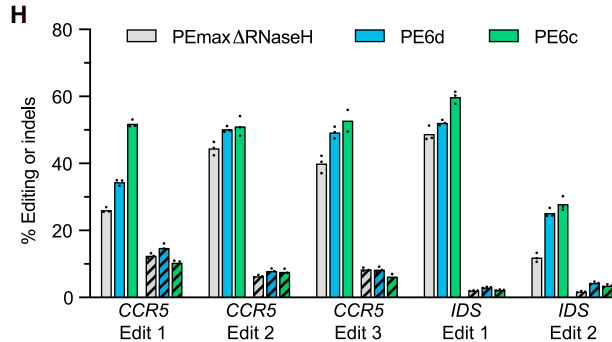
PE6b and PEmaxΔRNaseH were optimized for short templates, resulting in a 2.1-fold improvement in the edit-to-indel ratio (Fig. 4D), making them well-suited for high-precision SNP repair. The researchers applied these editors to Tay-Sachs disease model cells, achieving a 16–53% correction rate of the pathogenic mutation (Fig. 4H).
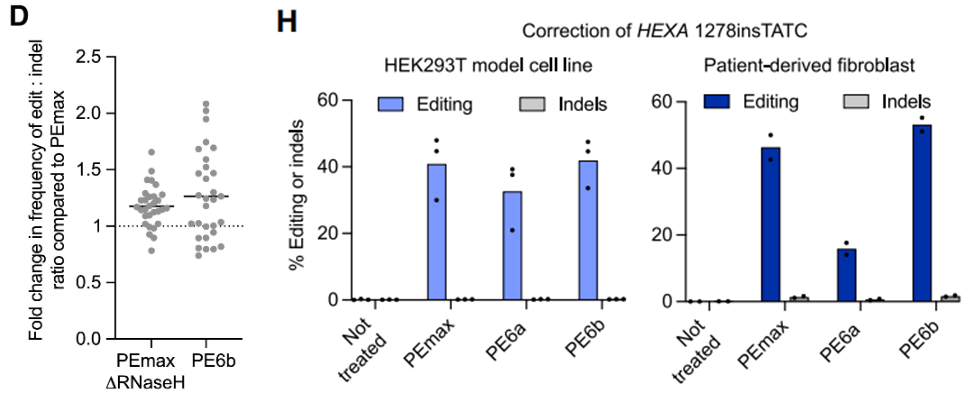
2. Long Fragment Knock-ins
PE6c/d achieved a 1.6-fold efficiency improvement in twinPE editing mediated by dual pegRNAs (Fig. 5H), enabling seamless insertion of fragments over 100 bp. This approach can be applied to scenarios such as recombinase recognition site installation and reporter gene knock-ins.
V. Conclusion and Outlook
This study successfully developed more compact and efficient Prime Editors (PE6) through phage-assisted evolution and protein engineering, representing a new breakthrough in gene editing technology. The PE6 series of Prime Editors demonstrate significant advantages in editing efficiency, editor size, and in vivo applications, opening new possibilities for future gene therapy.
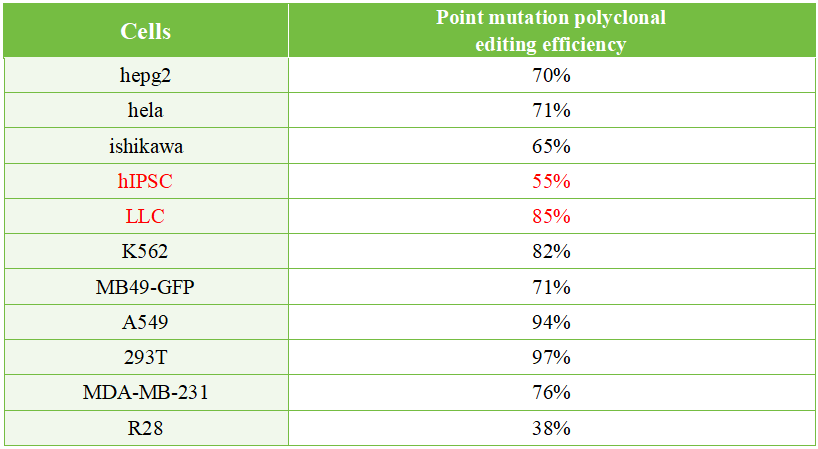
EDITGENE’s BINGO point mutation platform has provided gene point mutation services to thousands of customers. The cell pool editing efficiency reaches up to 95% in cell lines such as 293T and A549, and up to 80% in challenging-to-transfect and edit cell lines like LLC and K562.
Recent Blogs
- 1. [Literature Review] A New Hope for Progeria Treatment: CRISPR Gene Editing Uncovers Protein Synthesis Dysregulation and Novel Therapeutic Targets
- 2. [Literature Review] Unveiling T Cell Dynamics: CRISPR Library Screening and Single-Cell Transcriptomics Reveal Gene Regulation Networks
- 3. [Weekly News] From "Gene Scissors" to "RNA Erasers": A New Skill of the CRISPR-Cas System Has Been Unveiled
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com














Comment (4)