Alextina | Dec 28,2024
[Literature Review] The Flexibility of the Cas12a Domain Guides R-loop Formation and Forces RuvC Reset
The unique properties of CRISPR-Cas12a make it an ideal RNA-guided nucleic acid endonuclease for biotechnology and therapeutic applications.
Existing research has shown that Cas12a initiates R-loop formation by recognizing and binding to the protospacer adjacent motif (PAM) near the original spacer short palindromic repeat sequence, thereby identifying the target DNA. The formation of the R-loop triggers the activation of the Cas12a RuvC domain, which then cleaves the target DNA.
However, the specific steps and structural changes associated with Cas12a R-loop formation, the resetting mechanism of the RuvC lid, and the mechanisms maintaining Cas12a's specificity remain inadequately explained, and no detailed studies have described the flexibility of the Cas12a domains.
Scientists are eager for more in-depth research to gain further structural insights, which could advance CRISPR-Cas12a-based gene editing technology and biotechnology.
Recently, researchers from the University of Texas at Austin published their findings in Molecular Cell titled “Cas12a domain flexibility guides R-loop formation and forces RuvC resetting” This study, using cryo-EM technology, further explores Cas12a’s structure and function, providing structural insights that rationalize Cas12a's DNA targeting behavior and higher specificity.

Original Article Link: https://doi.org/10.1016/j.molcel.2024.06.007
I. Structure of Cas12a During the Cutting Process
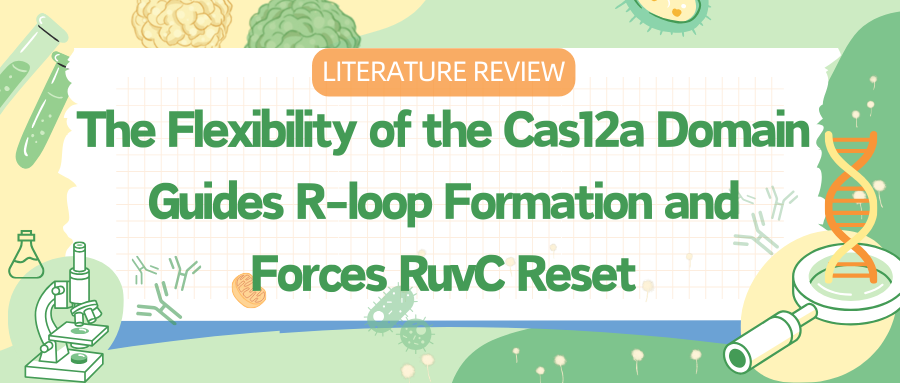
To capture various intermediates of WT Cas12a during R-loop formation, researchers incubated WT Cas12a with target DNA substrates of varying complementary sequence lengths to crRNA and obtained cryo-EM images. From these, they identified six different R-loop intermediate structures, with nominal resolutions ranging from 3.3 to 3.7 Å.
These structures of WT Cas12a observed in the cryo-EM results depict transient intermediate states of the R-loop, while also revealing several broadly observed findings. First, the number of decomposed R-loop base pairs does not always correspond to the extent of target complementarity.
For instance, the 8 bp target dataset produced two structures, 5-bp and 8-bp R-loops, and the 12 bp target dataset produced a new intermediate, a 10 bp R-loop. This suggests relative energy barriers in the R-loop formation process.
Secondly, the REC2 domain remains unresolved during intermediate R-loop propagation. Finally, during R-loop propagation, the distal DNA migrates from the front to the back of Cas12a, connecting the non-target strand (NTS) along the RuvC domain.
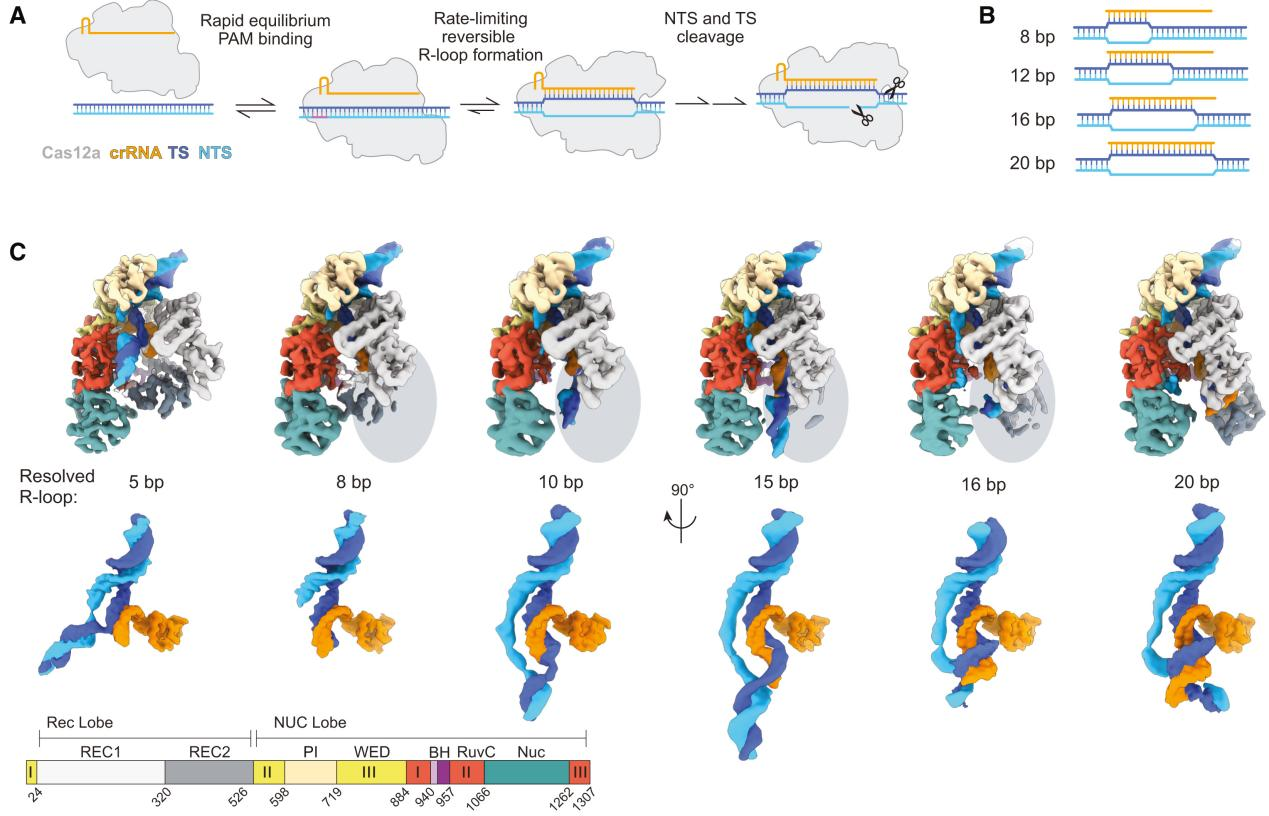
Figure 1 Cryo-EM Captured Intermediates of Cas12a R-loop
II. Cas12a Utilizes a 5 bp R-loop Seed Region for Target Recognition Activity
Similar to other RNA-guided effectors, Cas12a relies on a seed region within the R-loop to facilitate effective target recognition and mismatch discrimination.
In this study, the first structure of Cas12a in contact with the 5 bp seed region revealed that the first three nucleotides of the non-target strand (NTS) were captured by the Protospacer Adjacent Motif (PAM) interaction (PI) domain, while the REC1 domain moved outward to accommodate the formation of an A-form heteroduplex.
Most interactions were concentrated within the first few base pairs of the R-loop, formed by the Wedge (WED) and PI domains. The REC1 domain made few nonspecific stabilizing contacts with the R-loop at the seed terminus (K51, N175, R176).
The target strand (TS) and NTS rehybridized at the sixth base pair, located between the RuvC domain and the REC1 helix-loop-helix, guiding the distal DNA to exit the complex. The DNA at the R-loop-DNA junction was positioned on a bulky loop of the RuvC domain, with K1054 protruding into the minor groove of the DNA.
These results allowed the researchers to categorize structures with the same 5 bp R-loop and unresolved REC1 domain.
To explore the role of the RuvC loop in early R-loop structures, the researchers constructed and purified a loop deletion (LD) mutant and measured its effect on the cleavage of matched and mismatched targets.
The slightly reduced cleavage rate of seed-mismatched substrates in the LD mutant suggested that the loop contributes to stabilizing the 5 bp intermediate in the PAM-bound state of the WT enzyme.
The increased specificity observed in the LD mutant may result from destabilizing the early R-loop intermediate and increasing the R-loop collapse rate of both DNA substrates, leading to a measurable decrease in mismatch presence.
These results support the importance of the RuvC loop in stabilizing the seed region during R-loop formation.
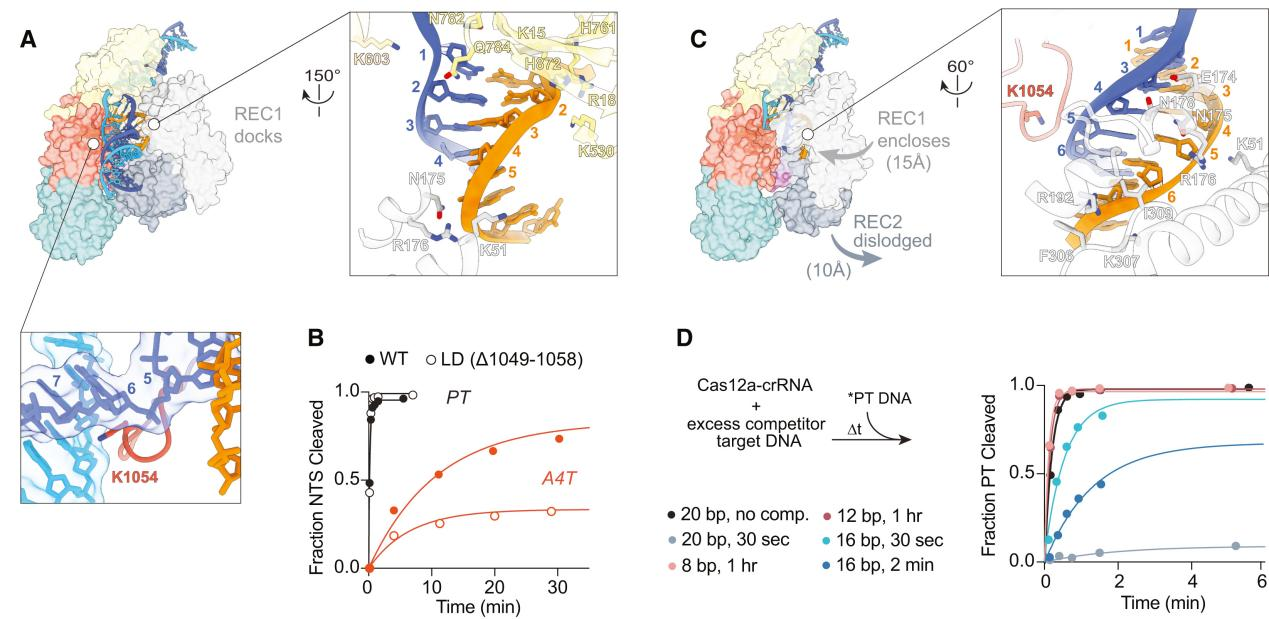
Figure 2 Cas12a Uses a 5 bp Seed to Rapidly Check DNA Complementarity
III. Rapid Dissociation Kinetics of Early and Mid R-loop Intermediates Facilitate Efficient Target Search
To functionally measure the stability of short R-loop intermediates, researchers tested their competitive inhibition of Cas12a targeting matched target DNA.
The results showed that 16 bp competitive targets exhibited preincubation time-dependent inhibition, indicating that these mid-R-loop intermediates can stably bind but are likely held in a reversible R-loop region.
The 20 bp competitors effectively inhibited all cleavage of protospacer-target (PT) DNA and demonstrated that Cas12a stably binds to matched targets within 30 seconds with minimal dissociation.
This rapid dissociation of R-loop intermediates may ensure that Cas12a is not sequestered on incorrect DNA sequences, enabling efficient search for the correct target. The rapid dissociation kinetics of these competitive reactions support that the early and mid intermediates observed in the cryo-EM data represent transient structures in the reversible R-loop formation process.
IV. REC2 Flexibility Accommodates R-loop Extension but Delays Contact
The conformational flexibility of the REC2 domain is one of the most notable changes observed in R-loop intermediate structures. The R-loop intermediates obtained by researchers indicate that the REC2 domain exhibits significant flexibility during R-loop formation. This flexibility allows for R-loop extension but delays contact with the Cas12a protein during the process.
This reveals that in the early stages of R-loop formation, the Cas12a protein does not stabilize intermediates, allowing for the reversibility of the R-loop and rapid dissociation from incorrect DNA sequences.
Additionally, cryo-EM observations showed dynamic changes in the REC2 domain and the bridging helix (BH) domain during intermediate R-loop formation. These changes are associated with the extension of the R-loop and the exposure of the active site.
In the 16 bp R-loop structure, REC2 begins to dock with the R-loop in the correct orientation but occupies the structure with low frequency due to a lack of sufficient stable contacts. As the R-loop extends, its shape changes, particularly at the distal end of the R-loop, affecting the docking of the REC2 domain and the stability of the BH domain.
These results suggest that the flexibility of the REC2 domain plays a crucial role in the DNA targeting and nuclease activation of Cas12a. It allows for the dynamic formation and extension of the R-loop while enhancing Cas12a’s DNA targeting specificity by delaying contact formation.
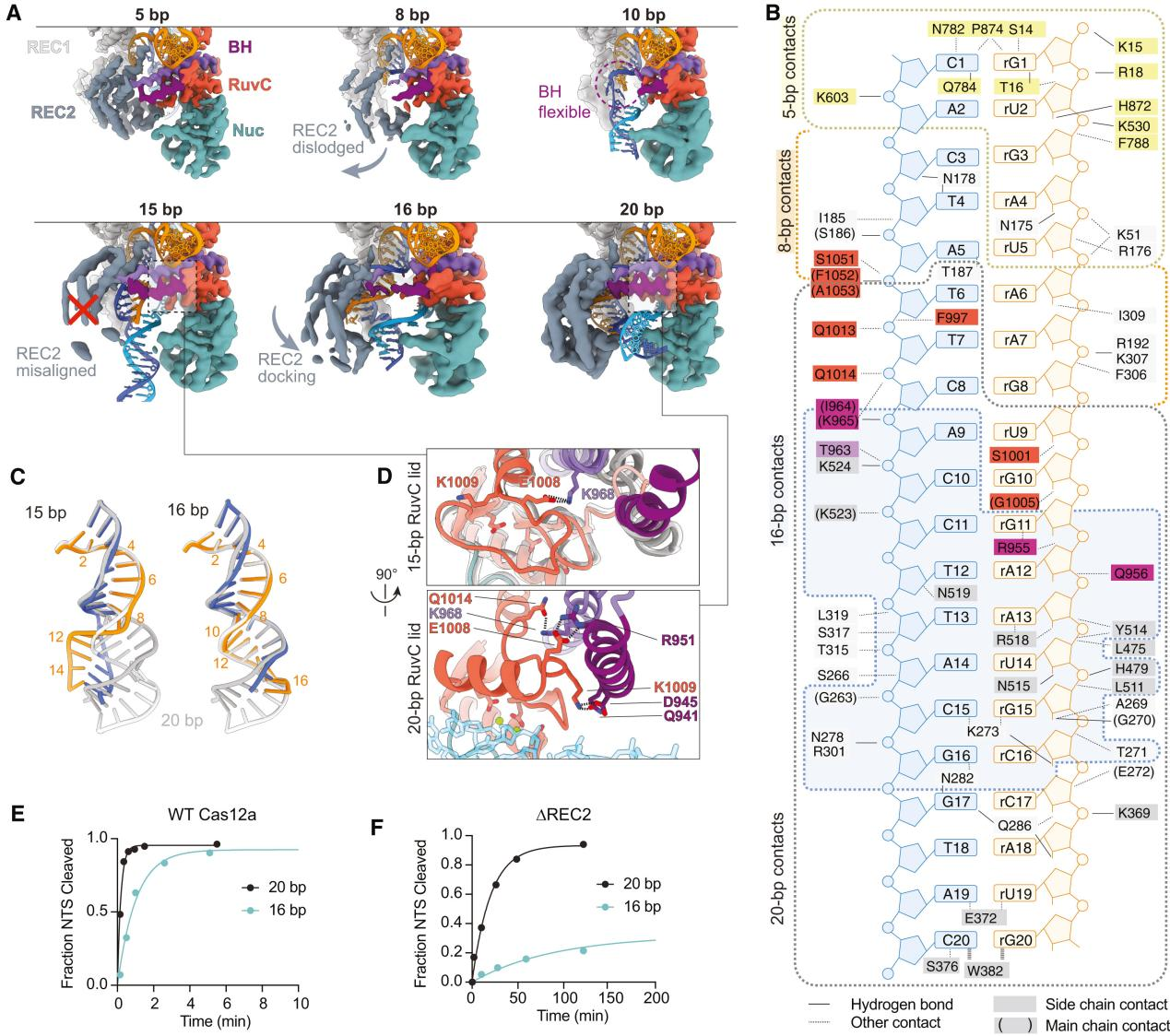
Figure 3 Characteristics of Intermediate R-loop Propagation Include REC2 Flexibility, Delayed R-loop Contact, and RuvC Activation
V. Late-stage REC2 Docking Promotes Catalysis
Previous reports have indicated that Cas12a requires at least a 15 bp R-loop to activate the RuvC nuclease for cleavage. The structural framework provided by the R-loops in this study explains how the minimum length R-loop leads to nuclease activation.
As the R-loop forms, the RuvC active site is blocked by the RuvC lid until a 15 bp R-loop structure is formed. The conformational rearrangement of the bridging helix (BH) establishes contact with the newly formed RuvC lid α-helix, exposing the active site.
Researchers proposed a mechanistic model where an R-loop of sufficient length to allow semi-stable REC2 docking achieves RuvC activation through BH contacts. However, the flexibility of REC2 and BH results in reduced efficiency.
To support this model, researchers tested the NTS cleavage of DNA substrates by Cas12a forming 16 or 20 bp R-loops. The results showed that the NTS cleavage rate of the 16 bp target was reduced by five times compared to the 20 bp R-loop.
Additionally, to further test the role of REC2 docking in RuvC activation through BH anchoring, researchers constructed a mutant lacking the REC2 domain (ΔREC2) for study. The results indicated that NTS cleavage rates for the 20 bp target were reduced by 65 times and for the 16 bp target by 83 times compared to WT Cas12a.
These data suggest that REC2 docking senses the R-loop length and transmits the signal to the RuvC domain through BH, thereby promoting activation.
VI. Non-Target Strand Preparation for Cleavage at the Active Site
Using cryo-EM technology, researchers captured the complete structure of Cas12a bound to a 20 bp R-loop to analyze the state of the non-target strand (NTS) poised for cleavage at the active site.
The findings revealed that during the final stages of R-loop formation and extension, the NTS is translocated to the RuvC nuclease domain, properly positioned within the active site, and prepared for cleavage.
The NTS is stabilized within the RuvC active site by a network of positively charged residues, including R912, K949, K1072, R1127, R1172, R1226, and N1295. Mutational analysis showed that the RuvC lid α-helix plays a crucial role in stabilizing the NTS, particularly through interactions between residues F999 and R1003 and the NTS.
To better understand the dynamics and role of the RuvC lid during the initial stages of NTS cleavage, researchers conducted molecular dynamics (MD) simulations using adaptive biasing molecular dynamics (ABMD) methods to study the transition of the lid from an unstructured loop to an α-helix.
Observing structural changes, they found that after NTS cleavage, the REC2 domain becomes highly dynamic, the Nuc domain gains flexibility, and the RuvC lid returns to an unstructured loop, closing the active site.
The target strand (TS) begins to move towards the active site, with the movements of the REC2 and Nuc domains aiding in the TS entry into the active site. The RuvC lid's α-helix also plays a role in TS cleavage.
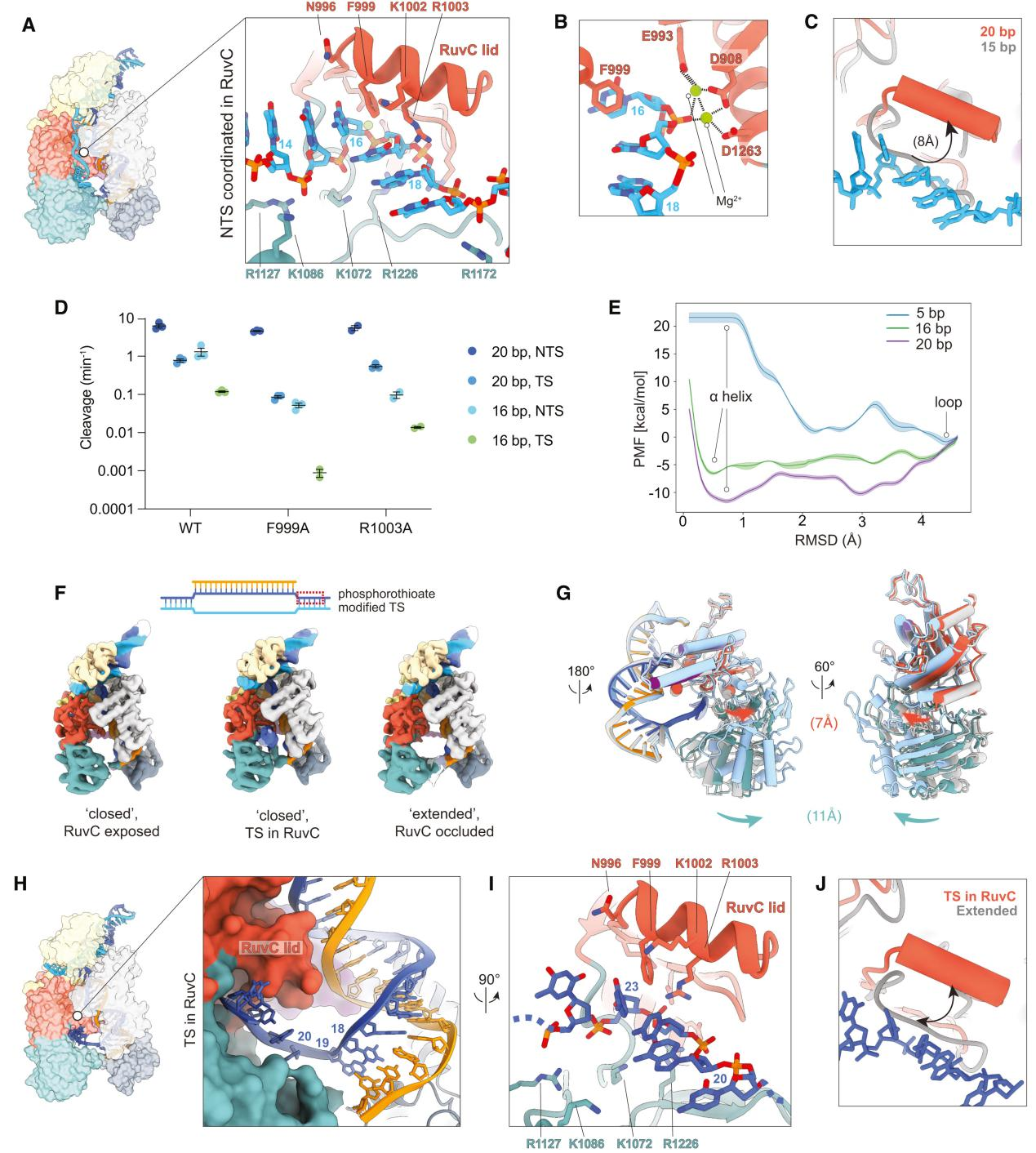
Figure 4 RuvC-mediated DNA cleavage by Cas12a
VII. Conformational Changes and Active Site Resetting in Cas12a Before Target Strand Cleavage
To understand the significant conformational changes in Cas12a before the cleavage of the target strand (TS) at the RuvC active site and how these changes lead to the resetting of the active site, researchers conducted further studies.
Firstly, they incubated Cas12a with a 20-bp target DNA modified with a phosphorothioate (ps) bond at potential TS cleavage sites. Through multiple rounds of 3D variability analysis, researchers captured significant domain flexibility in Cas12a during the RuvC-mediated TS cleavage process and captured the TS in the active site.
They found that after the cleavage of the non-target strand (NTS), the REC2 domain became highly dynamic, detaching from the R-loop and repositioning itself. The Nuc domain also exhibited flexibility as the TS approached the active site and moved together with the REC2 domain to help the TS enter the active site.
The RuvC lid underwent significant conformational changes before and after TS cleavage. During the approach of the TS to the active site, the movements of the REC2 and Nuc domains helped the TS enter and properly position itself within the RuvC active site.
Additionally, the researchers tested the TS cleavage efficiency of the F999A and R1003A mutants, finding that these mutations significantly reduced TS cleavage efficiency, especially for the 16-bp target. This indicates that phenylalanine (F999) in the RuvC lid is crucial for stabilizing the TS within the active site.
The results suggest that the RuvC active site needs to reset after NTS cleavage to prepare for the next TS cleavage. This resetting involves the coordinated movement of the REC2 and Nuc domains, as well as conformational changes in the RuvC lid.
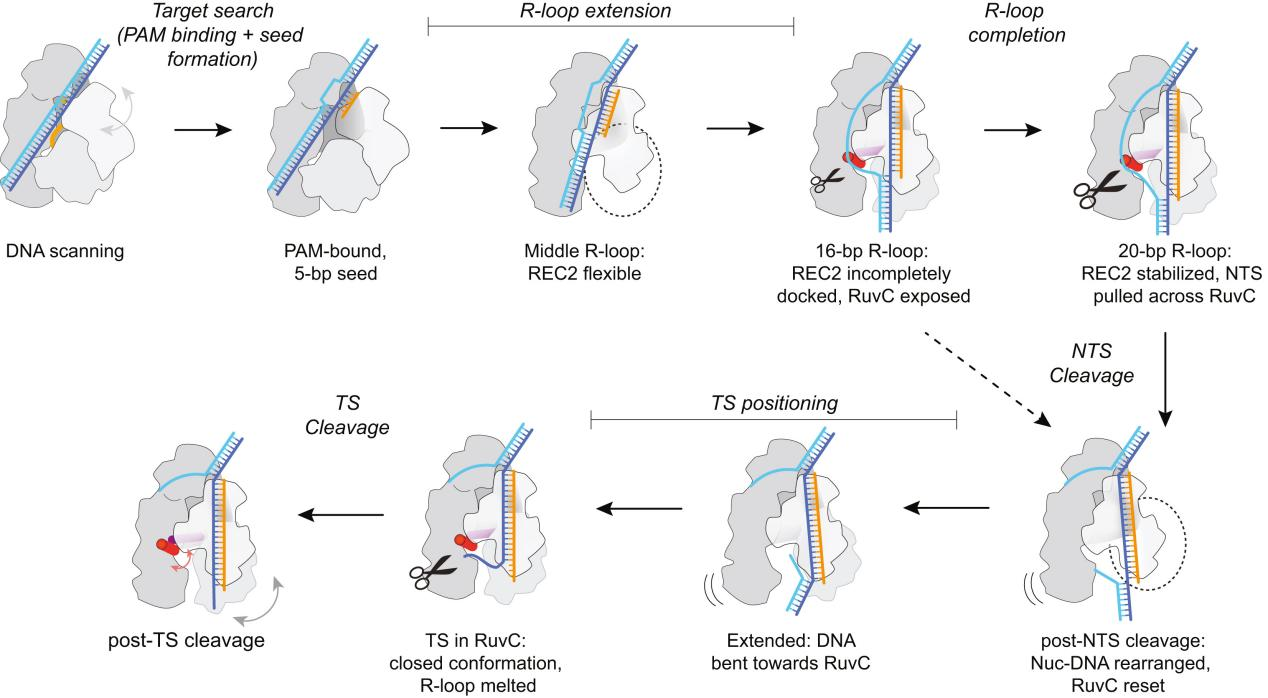
Figure 5 Structural Model of Cas12a R-Loop Formation and DNA Cleavage
In summary, researchers conducted an in-depth analysis of the structural changes in the Cas12a protein during R-loop formation and DNA cleavage using cryo-electron microscopy (cryo-EM) and other biochemical methods.
This study provides detailed insights into the molecular mechanisms of Cas12a in the CRISPR-Cas system for gene editing, particularly how it achieves high-specificity DNA targeting and cleavage through domain flexibility and conformational changes. The findings offer potential directions for further engineering and applications based on CRISPR-Cas12a.
EDITGENE boasts its own independently developed protein purification platform . Our Cas enzymes have shown significantly superior activity compared to NEB and other market enzymes through active testing, achieving the highest standards in purity, activity, and sensitivity.
Be on the look out for our isothermal amplification kit and CRISPR custom detection services , featuring low equipment requirements and short reaction times—Coming soon!
Recent blogs :
2. [Literature Review] Multiplex Detection Strategy of Biosensors Based on CRISPR-Cas System
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com


 Login
Login





![[Literature Review] The Flexibility of the Cas12a Domain Guides R-loop Formation and Forces RuvC Reset](/uploads/20241012/53c82bdd67704fe0e159246934f924ee.png)



Comment (4)