Knockout Cell Line
Knockout Cell Line
Service Details
| Cell Line | Tumor cells, Mmortalized cell line and iPSCs , etc. Konckout Cell Lines Bank |
|---|---|
| Service | Single-gene knockout / multi-genes knockout |
| Deliverables | Homozygous KO cell clones: ≥1 clone (2 vials per clone, 1 × 10^6 cells per vial) |
| Lead Time | 6 weeks |
| Price |
Price depends on KO cell lines and project difficulty |
Knockout Service Advantages
Efficient Guide RNA Design
High-Performance Cas9 Protein
Efficient Cell Transfection
Streamlined Cell Screening Solutions
Knockout Cell Line Services
| Frameshift Mutation | Guide RNA is targeted to an exon and the number of deletion bases is not a multiple of three. After knockout, a code-shifting mutation would cause gene knockout. |
|---|---|
| Large Fragment Deletion | Strategic Guide RNA design to enable the deletion of substantial gene segments. |
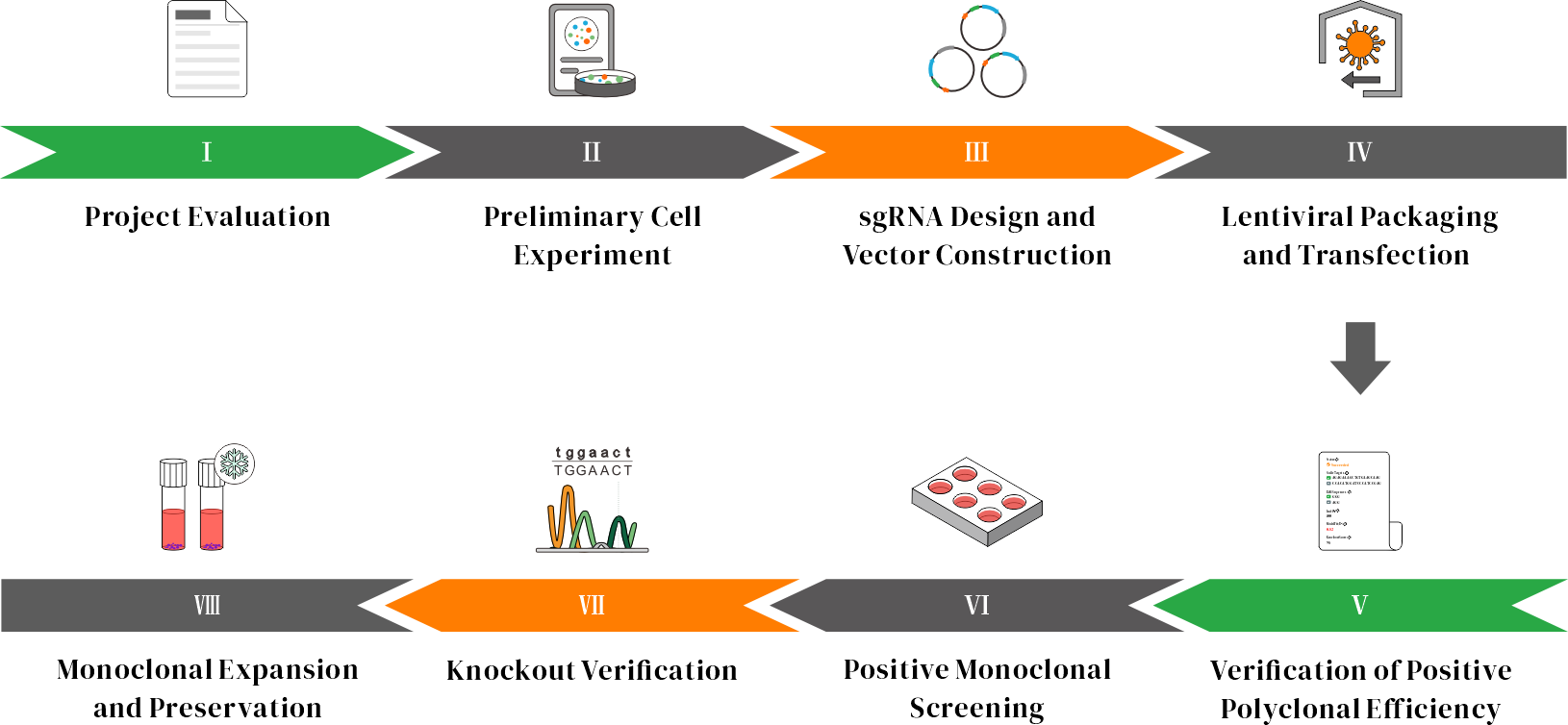
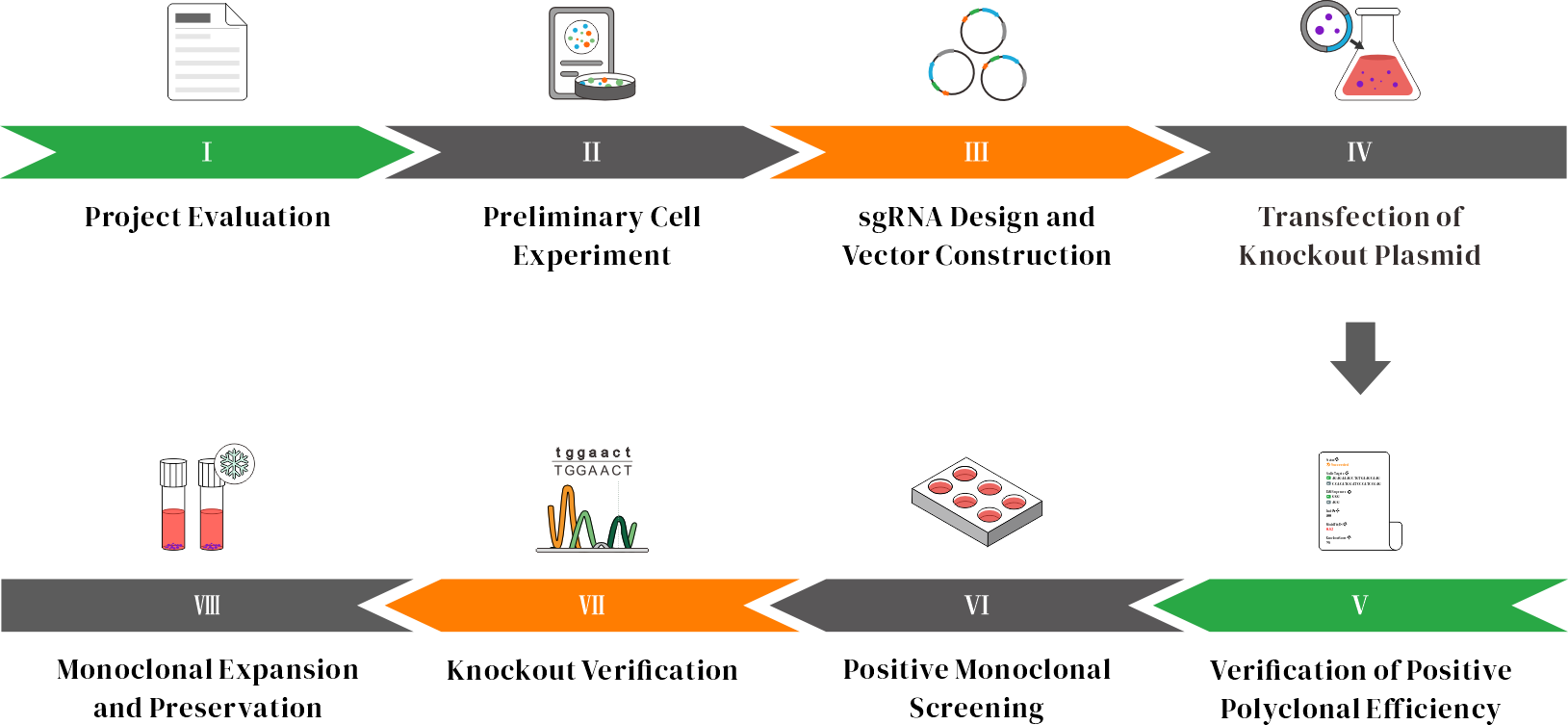
CRISPR Knockout Service Workflow
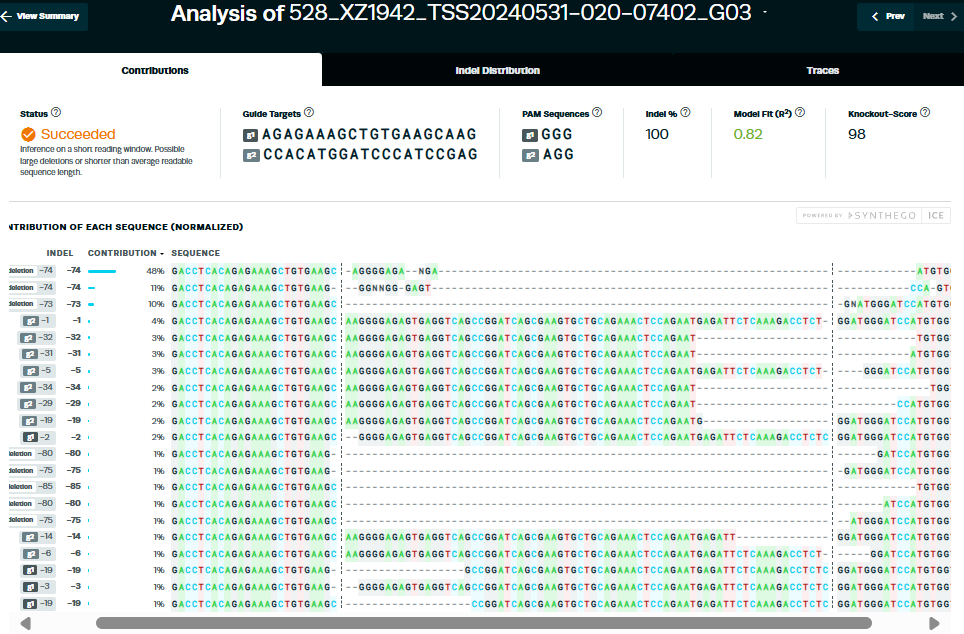
Gene Knockout Cell Service Case
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Genetic Reference Book
BRD4 gene knockout in skin squamous cell carcinoma (SCC) cells 
Recent studies show that the incidence of skin squamous cell carcinoma (SCC) is rapidly increasing, with this and other non-melanoma skin cancers leading to numerous deaths annually. Over 20% of the global population may develop skin cancer during their lifetime. BRD4 has been proposed as a potential oncogenic protein, but its role in skin SCC remains understudied.
Researchers used CRISPR/Cas9 to directly knockout the BRD4 gene in SCC cells. BRD4-knockout or silenced cells showed a significant reduction in proliferation, and the expression of oncogenes closely related to cell proliferation (e.g., cyclin D1, Bcl-2, MYC) was significantly decreased. In vivo experiments showed that BRD4 knockout significantly inhibited tumor growth in A431 cells in SCID mice. BRD4 knockout cells and its relevant findings suggest that BRD4 could be a therapeutic target in skin squamous cell carcinoma.
FGFR1 gene knockout in MDA-MB-231 breast cancer cell line 
The fibroblast growth factor (FGF)-fibroblast growth factor receptor (FGFR) signaling axis is a key mediator of interactions between the tumor stroma and cancer cells. FGFR1 activation through translocation, point mutations, or gene amplification can lead to cancer progression. In addition, G-protein-coupled estrogen receptor (GPER, GPR30) has been identified as a receptor mediating estrogen's role in various pathophysiological conditions.
To investigate how GPER mediates communication between cancer-associated fibroblasts (CAFs) and breast cancer cells through the FGF2/FGFR1 signaling axis, researchers used CRISPR/Cas9 gene editing to knockout FGFR1 in the MDA-MB-231 breast cancer cell line. Conditioned media (CM) from estrogen-stimulated CAFs induced the expression of connective tissue growth factor (CTGF) in FGFR1 wild-type (WT) MDA-MB-231 cells and promoted migration and invasion via the FGFR1-ERK1/2-AKT signaling pathway. However, this effect was significantly reduced or abolished in FGFR1-knockout cells. FGFR1 knockout cells have revealed a novel role of GPER in regulating FGF2 expression within the tumor microenvironment. The study confirmed that FGFR1 gene amplification is closely associated with overall survival rates in breast cancer patients, suggesting that FGFR1 could serve as a potential therapeutic target in breast cancer treatment. Furthermore, it elucidates the paracrine activation between cancer-associated fibroblasts (CAFs) and breast cancer cells, providing a theoretical foundation for the development of new therapeutic strategies.
BRCA1 gene knockout in Swan71 cell line 
In early placental development, tumor suppressor genes and oncogenes work together to regulate cell proliferation and differentiation. BRCA1 is a well-known tumor suppressor gene that forms a complex with ZNF350 and CtIP to bind to the promoter region of the HMGA2 gene, preventing its transcription. This regulation has been studied in cancer cells but less so in placental cells.
Researchers used the CRISPR-Cas9 system to knockout the BRCA1 gene in the Swan71 cell line, generating BRCA1-knockout cells. Lentiviral particles with miR-182 overexpression were used to overexpress miR-182 in Swan71 cells. The results showed that BRCA1-knockout cells had significantly higher HMGA2 mRNA and protein levels compared to wild-type cells. miR-182 overexpression led to a decrease in BRCA1 protein levels and an increase in HMGA2 protein levels. BRCA Knockout Cell Lines and its relevant findings demonstrate that BRCA1 plays an important role in regulating HMGA2 levels in trophoblast cells and may be involved in placental development and function by influencing apoptosis, providing new insights into BRCA1's role in placental development.
ATM gene knockout in NGP and CHP-134 NB cell lines 
Neuroblastoma (NB) is a common pediatric solid tumor characterized by high clinical and prognostic heterogeneity. Despite multiple treatment strategies, tumors in high-risk NB patients exhibit resistance to standard therapies and may progress to metastasis. ATM gene is involved in DNA damage response, and heterozygous deletions or hemizygous mutations of the ATM gene located on chromosome 11q are mutually exclusive in NB tumors. While ATM knockdown has been shown to promote tumor formation in NB cell lines in vitro and in vivo, the connection between ATM and tumor formation or cancer invasiveness remains unclear.
Researchers used CRISPR/Cas9 technology to knockout the ATM gene in NGP and CHP-134 NB cell lines, analyzing cell proliferation and colony formation capabilities, and protein expression related to DNA repair pathways through Western blot. In ATM-knockout cells, stable transfection of FANCD2 expression plasmids was used to overexpress FANCD2. Immunofluorescence microscopy was employed to determine protein expression. The results showed that ATM depletion leads to a decrease in FANCD2 protein levels, and ATM-knockout cells are more sensitive to the PARP inhibitor. Reintroduction of FANCD2 in ATM-knockout cells restored cell proliferation capacity. ATM knockout cells and its relevant findings reveal the role of ATM haploinsufficiency in neuroblastoma and illustrate how ATM inactivation enhances NB cell sensitivity to PARP inhibitor, which is significant for treating high-risk NB patients with ATM gene dosage and cancer progression issues.


 Login
Login