 Point Mutation Cell Line
Point Mutation Cell Line
Service Details
| Cell Type |
Various cell types including tumor cells, epithelial cells, and stem cells. Click to view the full list of cell lines |
|---|---|
| Service Types | Model construction / Efficiency validation for therapeutic applications / DNA sourcing |
| Delivery Standard | Gene point mutation monoclonal cell line ≥ 1 clone (2 vials per clone, 1×10^6 cells per vial) |
| Lead Time | Projects as short as 10 weeks |
| Price |
|
EDI-Service Advantages 1
Efficient Editing System
Optimized pegRNA Design
Enhanced Cas9n-RT Enzyme
Advanced Transfection System
Streamlined Monoclonal Screening
Experienced Team
Service Advantages 2
EDITGENE has recently achieved a major breakthrough with its Bingo™ platform Prime Editing technology, officially launching Bingo™ PE7, update from last generation Bingo™ PE5.
Prime Editing 7 Technology Highlights
1) 20% Higher Success Rate – Achieve Results with Confidence!
Thanks to EDITGENE’s advancements, PE7 has optimized the binding activity between editing proteins and RNA, boosting point mutation success rates by 20% compared to PE5. This isn’t just a number—each experiment is now more precise and reliable. Higher success rates mean less stress and lower costs, allowing researchers to focus on groundbreaking discoveries.
2) 4.7x Increased Efficiency at Challenging Sites – Turning Challenges into Opportunities!
PE7 brings a major performance boost, reaching up to 86% editing efficiency at easy-to-edit sites (where PE5’s efficiency was above 20%). The real strength of PE7 is its power to tackle tough sites: for challenging targets where PE5’s efficiency was under 20%, PE7 raises efficiency by 1.76x to 4.71x! With PE7, previously difficult editing tasks become achievable, opening doors to new research possibilities.
The fold increase in editing efficiency of PE7 compared to PE5
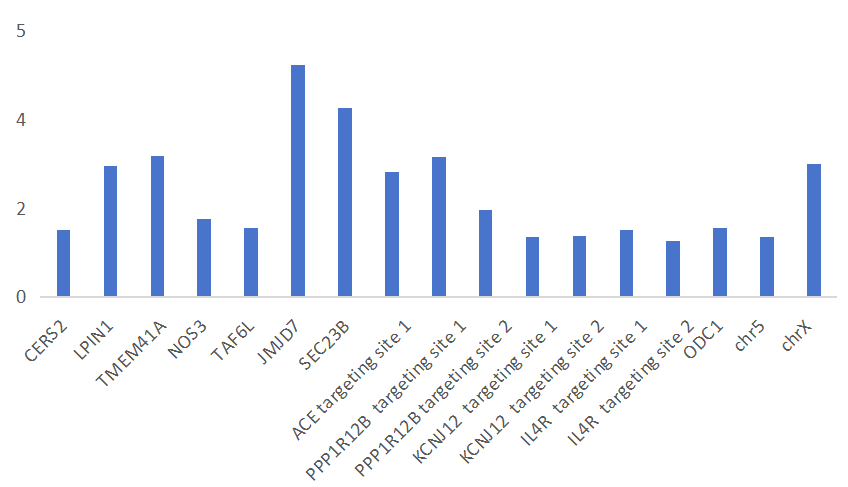
Sites where editing failed via PE5, but were successfully edited via PE7
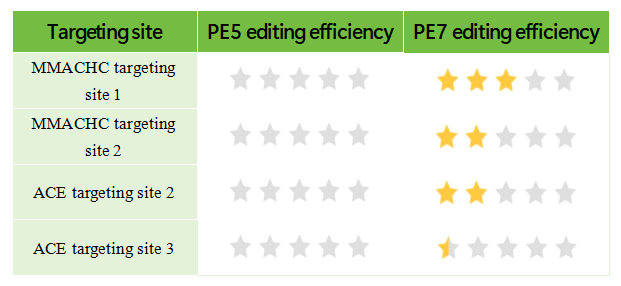
Note: 1 grade<10%, 2 grade20%-30%, 3 grade30%-40%,4 grade50%-60%,5 grade 70%-100%
Improvement in editing efficiency of PE7 compared to PE5
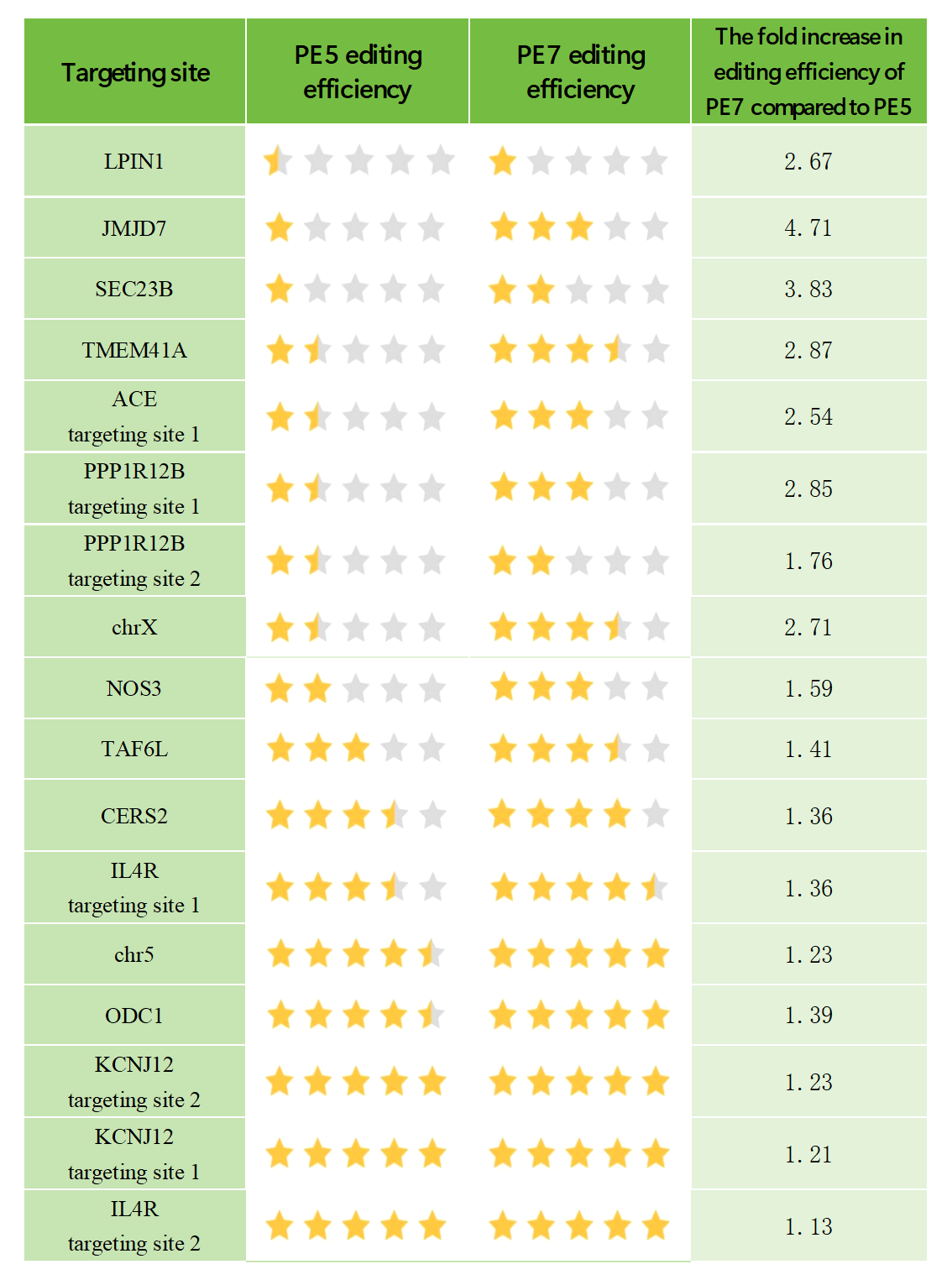
Note: 1 grade<10%, 2 grade20%-30%, 3 grade30%-40%,4 grade50%-60%,5 grade 70%
EDI-Service Types
| 1 | Construction of point mutation cell line models |
|---|---|
| 2 | pegRNA efficiency validation for therapeutic applications |
| 3 | Biological source material for standard DNA in in vitro diagnostics (IVD) |
EDI-Service Workflow
Case Study
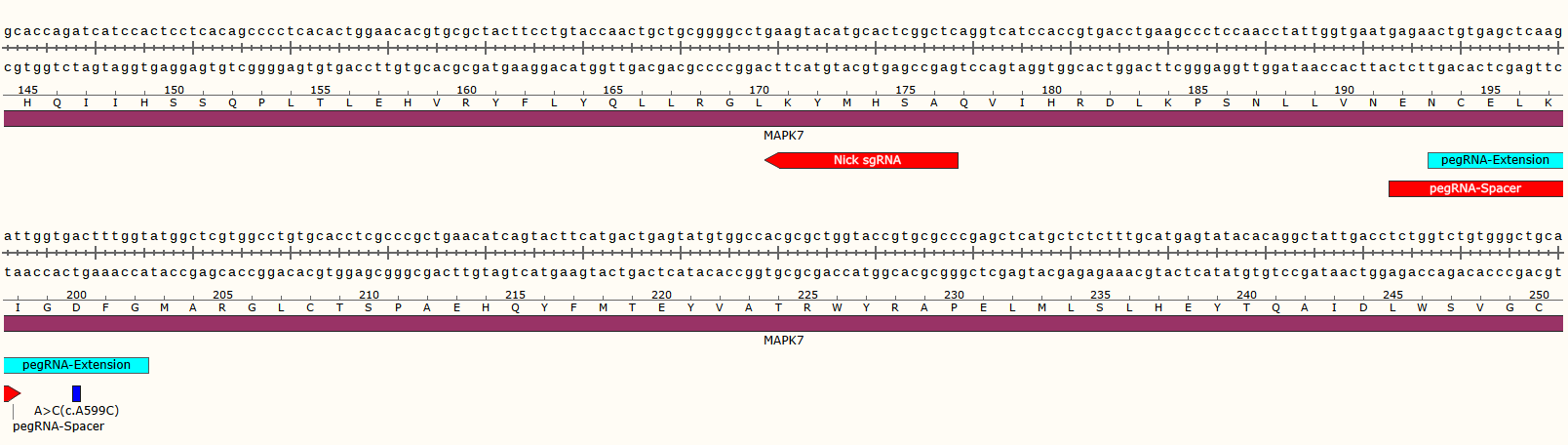
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Genetic Reference Book
PE7 editing system developed based on LA as a prime editing enhancer 
Many studies focus on enhancing the performance of the Prime editor to increase its editing efficiency; however, the understanding of how Prime editing operates within cellular environments and how interactions with the cellular environment affect editing outcomes remains limited. Researchers sought to identify other cellular determinants affecting Prime editor efficiency and conducted related studies.
Through genome-scale CRISPR interference (CRISPRi) screening, researchers discovered a key Prime editing enhancer—La, a small RNA-binding exoribonuclease protector protein. The La protein binds to pegRNAs’ 3' polyU sequence through its N-terminal domain, enhancing Prime editing. This effect of the La protein is effective for different editing types (substitutions, insertions, deletions) and various cell types. Based on this finding, researchers developed a new Prime editor protein (PE7), in which the La RNA-binding N-terminal domain is fused to the PEmax editor. PE7 significantly improves Prime editing efficiency with expressed pegRNAs, engineered pegRNAs (epegRNAs), and synthetic pegRNAs optimized for La binding.
Cryo-EM study of Prime editor complex to facilitate iterative upgrades to prime editing 
The molecular mechanism of how the Prime editor recognizes pegRNA and interacts with target DNA remains unclear, limiting the understanding and optimization of the prime editing process. To address this, researchers determined the cryo-electron microscopy (cryo-EM) structure of the Prime editor in various states, providing a structural framework for understanding this innovative genome engineering system.
Using cryo-EM technology, researchers analyzed the Prime editor complex’s structure in different states, including pre-initiation, initiation, extension, and termination, successfully obtaining high-resolution structures that reveal dynamic changes in the reverse transcription guidance process. Based on structural information, researchers designed pegRNA and Prime editor variants, truncating and fusing M-MLV RT to create a smaller Prime editor variant (PECO-Mini) that maintained editing efficiency while increasing AAV vector titers and prime editing efficiency. Activity testing results showed that the engineered Prime editor variant has comparable activity to the original Prime editor in vitro. This study reveals the structural characteristics of the Prime editor complex in various working states, providing key information for understanding its molecular mechanism.
Increasing Prime editing efficiency via truncated RT enzyme with AAV delivery 
Prime editing is a novel CRISPR-based genome-editing technology that does not require double-strand DNA breaks or exogenous donor template DNA, showing great potential in biomedical research and gene therapy. Despite its versatility and precision, prime editing efficiency varies across different editing types, target sites, and cell types. Therefore, to expand its applications, there is a need to improve prime editing efficiency.
Researchers screened 11 different RT variants, optimized with GenScript algorithm for human codons, which increased PE protein expression levels by 1.4-fold. By deleting the RNase H domain and further shortening the RT sequence, they created multiple truncated PECO variants, reducing the Prime editor length by 621 bp without compromising editing efficiency. To enable efficient dual-AAV delivery of PE, the team constructed a split PE system based on different Cas9 cleavage sites and inteins, identifying cleavage sites Rma 573-574 and 674-675. When these sites were paired with Rma intein, they significantly increased AAV vector titers and Prime editor efficiency. Through engineering and optimization, researchers successfully enhanced Prime editing efficiency and resolved AAV size constraints, providing a more efficient tool for future gene therapy and biomedical research.
Iterative upgrades to prime editing technology by studying Prime editor’s mechanism of action 
This article elucidates the structural basis of pegRNA-guided reverse transcription in prime editing technology. High-resolution structural analysis revealed the three-dimensional conformation of key proteins in the Prime Editor, including Cas9 and reverse transcriptase, and their interaction with pegRNA. It details how pegRNA guides the Prime Editor to recognize specific DNA sequences and perform reverse transcription, inserting a predetermined gene sequence into the target site. The study also discusses the roles of key amino acid residues in pegRNA binding and reverse transcription, providing insight into the precise mechanisms of these molecular interactions. This finding not only deepens our understanding of the Prime Editing mechanism but also offers valuable structural information for optimizing and improving this technology, advancing its application in gene therapy and other fields.
Point mutation of the HBBS gene in hematopoietic stem and progenitor cells (HSPCs) 
Sickle-cell disease (SCD) is an autosomal recessive genetic disorder caused by an A·T to T·A point mutation in the β-globin gene (HBB). Currently, the only FDA-approved cure for SCD is allogeneic hematopoietic stem cell transplantation; however, most patients lack ideal donors, and this procedure can lead to severe toxicity. Correcting a patient’s own hematopoietic stem cells (HSCs) can bypass immune complications and eliminate the need for matched donors. Clinical trials are ongoing to correct the SCD mutation using Cas9 nuclease-initiated homology-directed repair (HDR) and adeno-associated virus type 6 (AAV6)-delivered DNA templates.
In this article, researchers applied an optimized prime editing system to correct HSPCs from SCD patients ex vivo. By electroporating PEmax mRNA along with synthetic epegRNA and nicking sgRNA, the SCD allele (HBBS) was successfully corrected back to the wild-type (HBBA), with correction rates between 15% and 41%. Subsequently, edited HSPCs were transplanted into immunodeficient mice, and 7 weeks later, edited HSPCs maintained HBBA levels in the bone marrow, with engraftment rates, hematopoietic differentiation, and lineage maturation similar to unedited healthy donor HSPCs. Therapeutic evaluation revealed that, post-transplant, 42% of red blood cell precursors and reticulocytes expressed HBBA, exceeding the predicted therapeutic benefit level. Gene-specific analyses also confirmed high DNA specificity of the prime editing system. The study demonstrates the potential of prime editing in SCD treatment, showing its effectiveness in improving therapeutic outcomes while reducing off-target editing risks.
Point mutation of multiple Type 2 Diabetes (T2D) risk SNVs in iPS cells 
With advancements in human molecular genetics and genomics, thousands of gene loci associated with common disease risks have been identified, often containing multiple candidate variants. However, most disease-associated variants are non-coding, making it challenging to elucidate the molecular mechanisms underlying these variants. The development of CRISPR technology provides a more precise approach for targeted genome editing, particularly prime editing, which can mediate nearly any single-nucleotide substitution. Consequently, many researchers hope to apply prime editing to study the functional relevance of specific gene variants in pluripotent stem cells, enabling the generation of isogenic cell lines to control genetic background while assessing the dose effect of causal alleles.
In this article, researchers developed an efficient CRISPR prime editing technology to generate cell lines carrying heterozygous or homozygous alleles in induced pluripotent stem cells (iPSCs) and optimized the prime editing technique. Six single-nucleotide variants (SNVs) associated with Type 2 Diabetes (T2D) risk were selected for editing, and iPSCs derived from human donors with different genetic backgrounds were edited at each site. The researchers successfully generated 27 edited iPSC clones covering 6 SNVs associated with T2D or congenital hyperinsulinemia (CHI) and found that prime editing was more efficient in iPSCs than in HEK293T cells. Overall, this study demonstrates the potential of prime editing for generating iPSCs with specific genetic backgrounds, providing a powerful tool to study the effects of specific genetic variants on disease.


 Login
Login



