 Knock-In Vector Construction
Knock-In Vector Construction
Service Details
| Service Types | Fluorescent Protein Knock-in Vector / Tag Protein Knock-in Vector / Specific DNA Fragment Knock-in Vector |
|---|---|
| Delivery Standards | 1.Plasmid map2.Plasmid sequencing results3.Plasmid amplification instructions 4.Plasmids (sgRNA-Cas9 editing plasmid and Donor plasmid) |
| Timeline/Pricing |
|
Service Advantages
Long Insert Fragments
High-Activity Cas9 Plasmids
Optimized sgRNA
Experienced Team
Service Types
| Fluorescent Protein Knock-in Vectors | EGFP, Luc, mCherry, etc. |
|---|---|
| Tag Protein Knock-in Vectors | Flag, HA, Myc, HiBiT, etc. |
| Vectors for Inserting Specific DNA Fragments into Target Genomic Loci | / |
| Vectors for Inserting Specific DNA Fragments into Genomic Safe Harbor Sites | / |
Plasmid Map
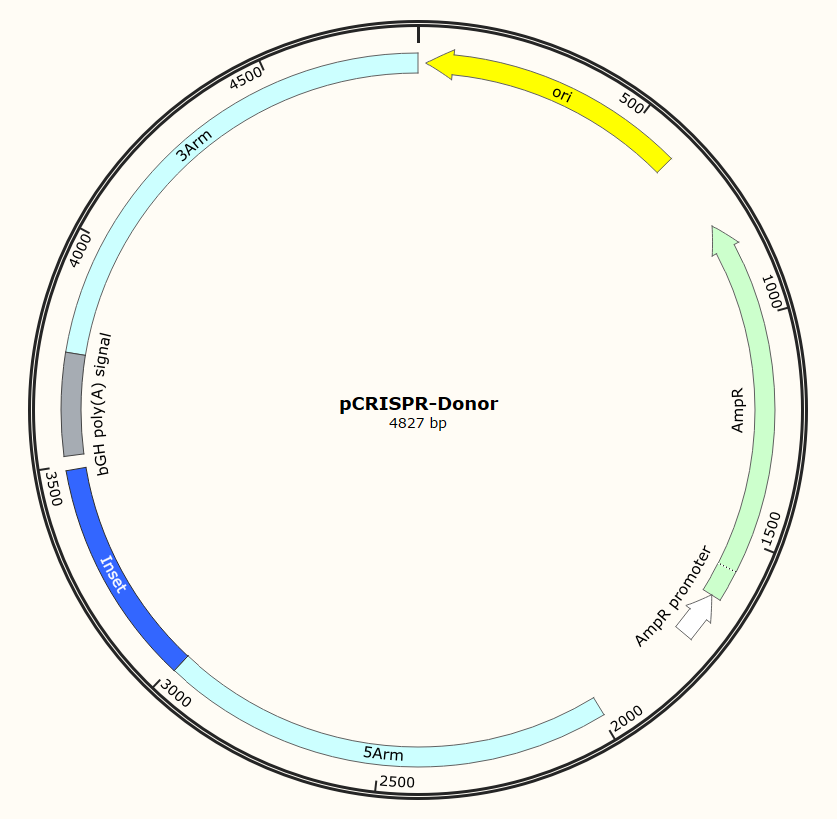
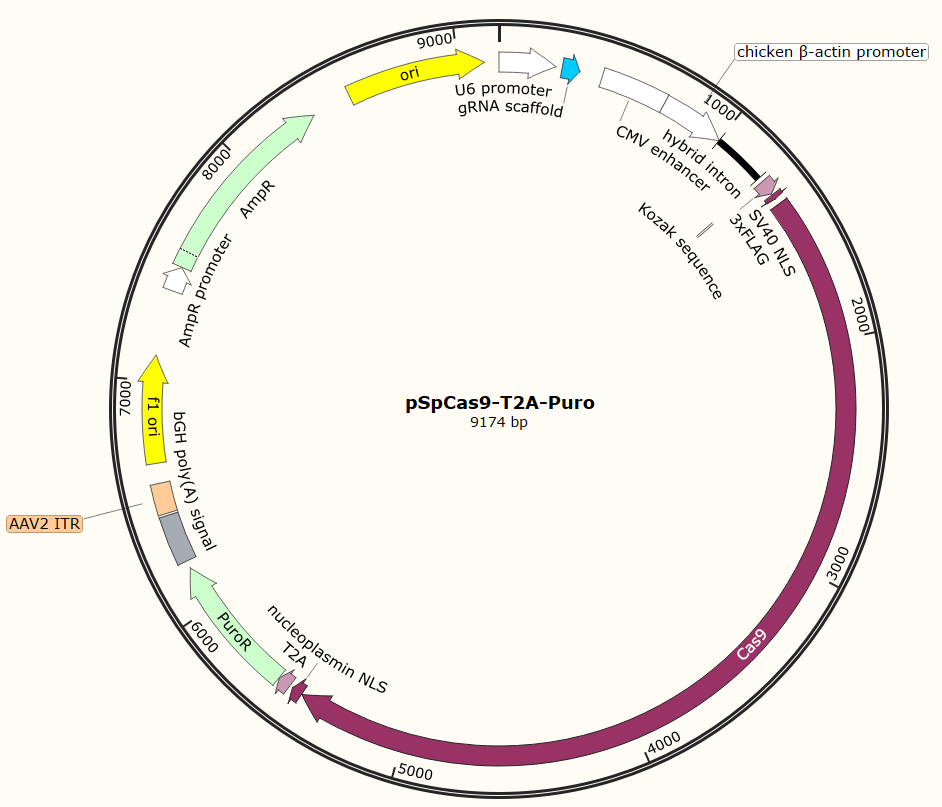
Advantage and Characteristic

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy

Optimazied Strategy
Genetic Reference Book
Insertion of tdTomato Downstream of the Ins2 Promoter in C57BL/6J Mice 
Using CRISPR/Cas9 gene editing technology, researchers constructed a transgenic mouse model to express a specific fluorescent protein in pancreatic β-cells. In C57BL/6J mice, the tdTomato transgene was inserted downstream of the Ins2 promoter. The researchers designed an Ins2-specific single-guide RNA targeting exon 2 for the CRISPR/Cas9 system and constructed a donor vector. The Cas9, single-guide RNA, and donor vector were then injected into mouse fertilized eggs in vitro, and these fertilized eggs were implanted into pseudopregnant mice. Heterozygous mating produced homozygous mice, and genotype identification, in vivo imaging, and frozen sections confirmed the knock-in effect. Six F0 mice and stably inherited Ins2-IRES-tdTomato F1 were obtained. Genome sequencing results showed no changes in the Ins2 exon compared to the control group; only the base sequence of tdTomato was added without base mutations. However, no expression of red fluorescent protein (RFP) was observed in vivo imaging and frozen sections, indicating low expression of the tdTomato protein and fluorescence intensity not reaching the detection threshold. In CRISPR/Cas9 technology, the IRES-linked foreign fragment may affect the transcription levels of the upstream gene, leading to low expression levels of the downstream gene and affecting the insertion effect.


 Login
Login



