[Customer Publication] Methylation “Fingerprint” Detection May Transform Early Cancer Screening
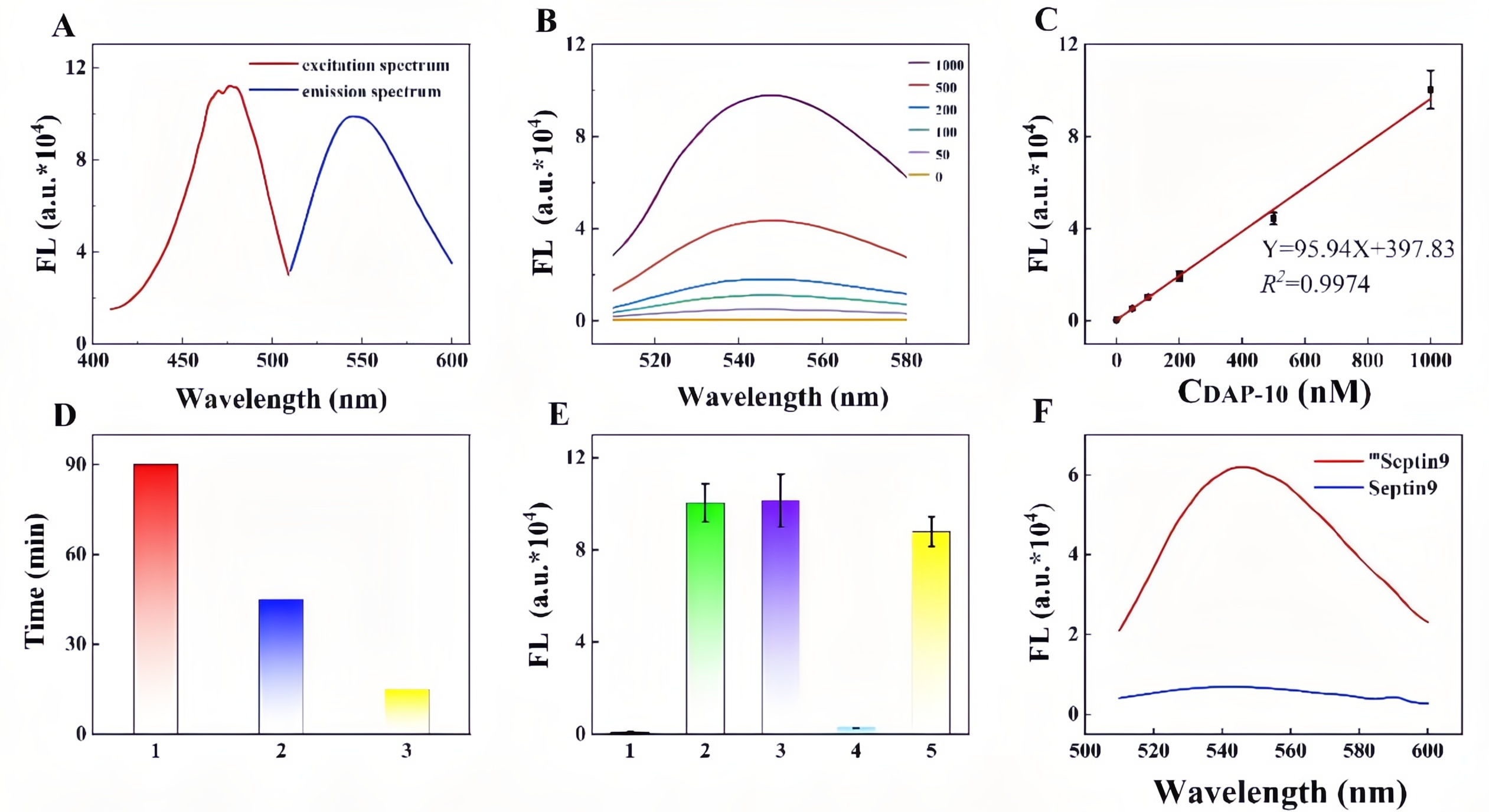
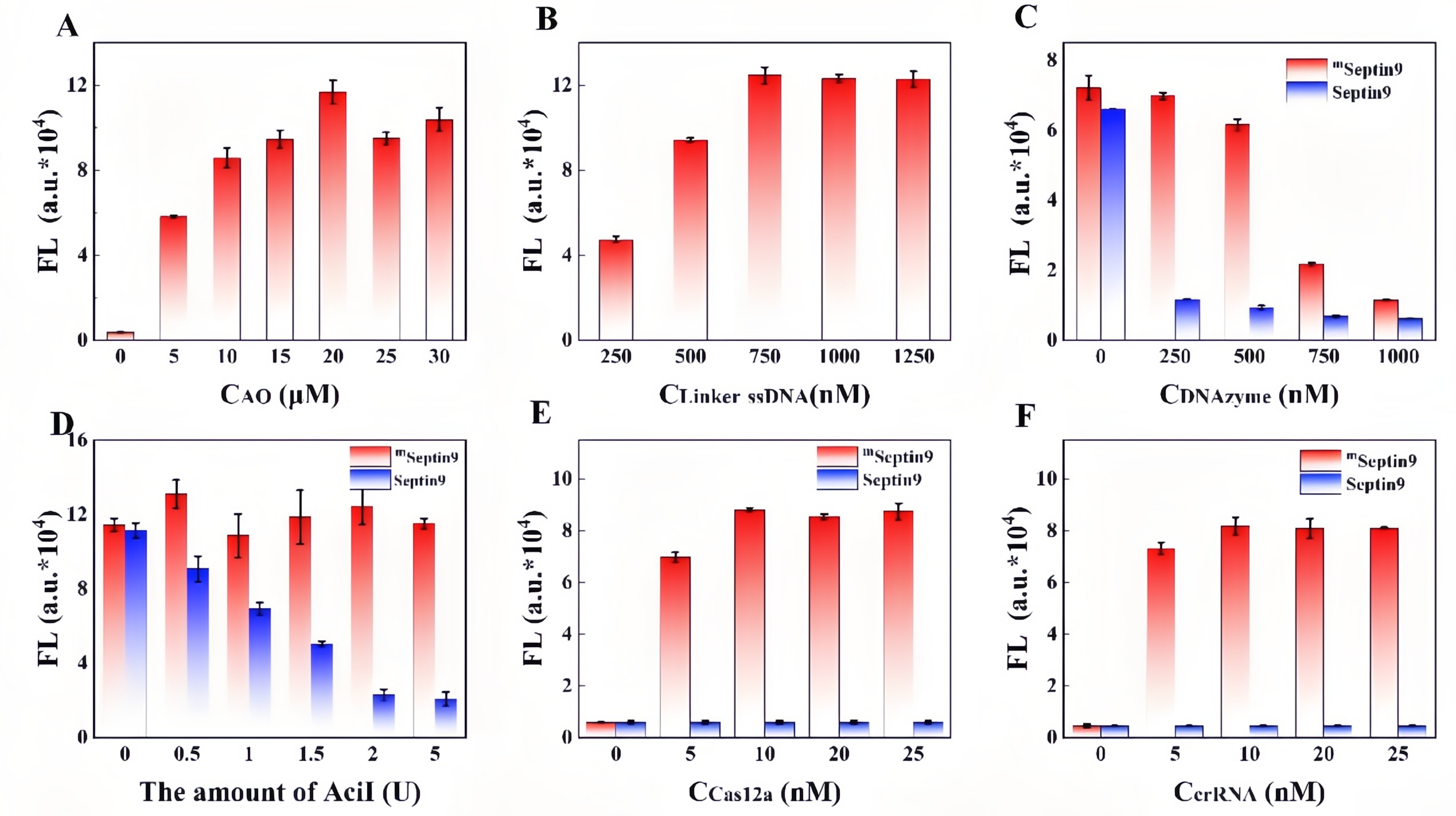
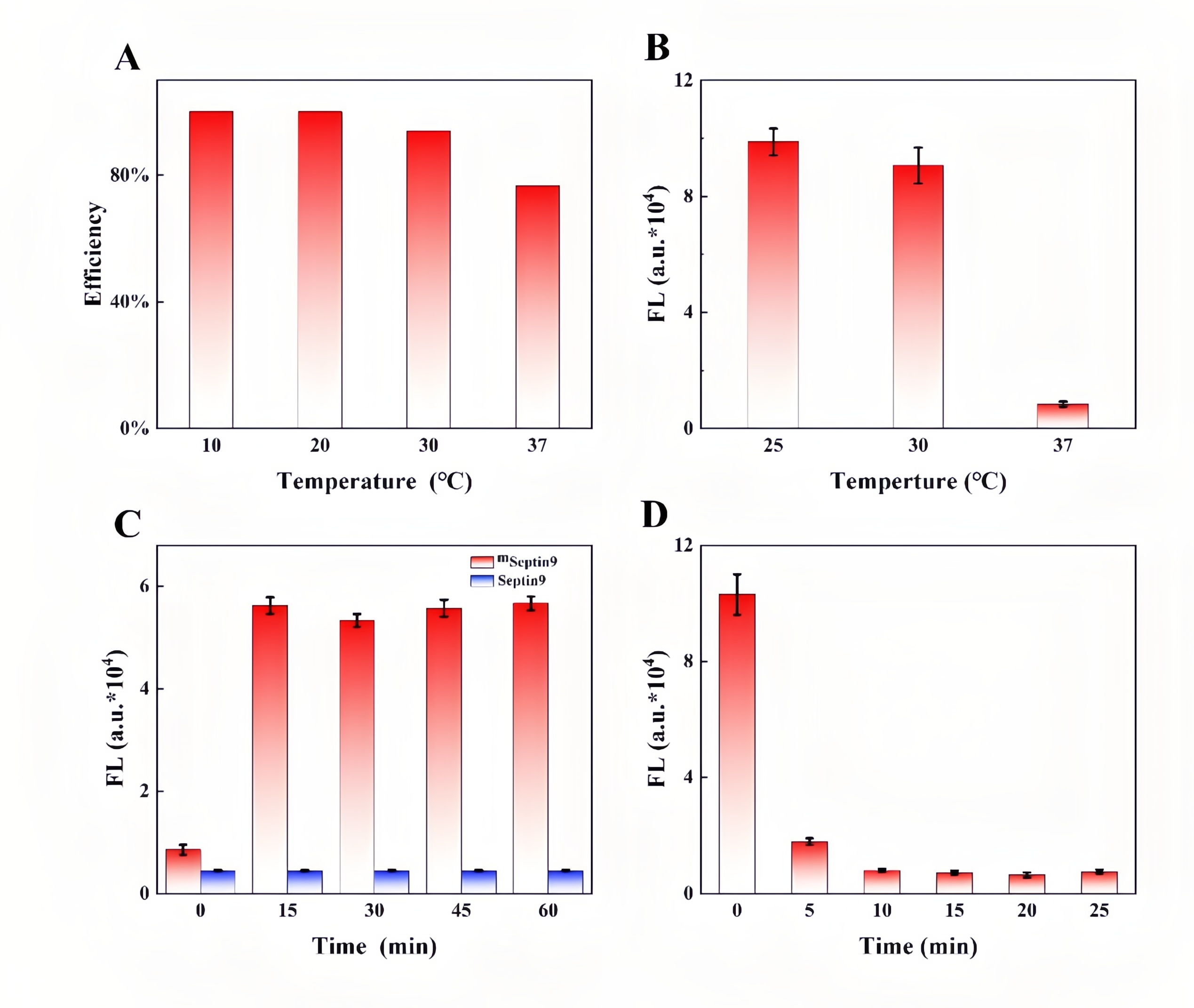
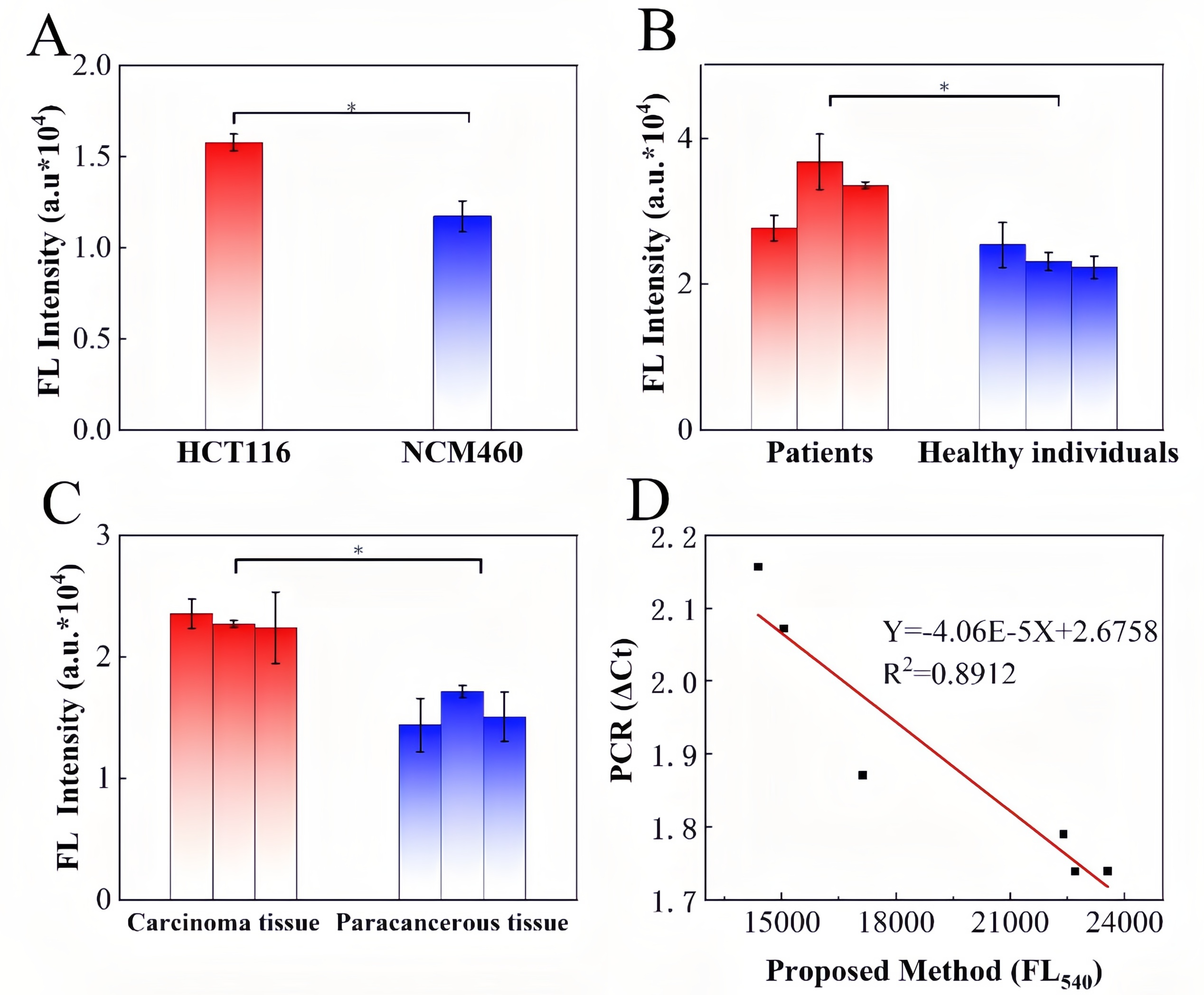
Colorectal cancer (CRC) is a highly prevalent malignancy worldwide, and early screening can significantly reduce mortality. While the FDA-approved Septin9 methylation blood test enables non-invasive screening, it relies on complex probes and expensive equipment, highlighting the need for simpler, cost-effective alternatives. Recently, the team led by Prof. Songcheng Yu at Zhengzhou University published a paper in Biosensors and Bioelectronics entitled “CRISPR/Cas12a-mediated DNAzyme/split-aptamer cascade for label-free detection of site-specific DNA methylation.” Their work addresses the limitations of conventional Septin9 methylation assays, including reliance on costly fluorescent probes and cumbersome workflows, by developing a CRISPR-Cas12a-mediated DNAzyme/split-aptamer cascade platform. The principle of the method is as follows: methylation-sensitive enzyme AciI first digests unmethylated sequences, leaving only methylated Septin9 fragments to activate Cas12a. The activated Cas12a then trans-cleaves the DNAzyme, preventing it from hydrolyzing the linker. This allows the split DAP-10 aptamer to reassemble and light up Auramine O fluorescence, producing a signal that directly correlates with methylation levels. EDITGENE was proud to support this research by providing high-performance Cas12a proteins, contributing to the successful outcome of the study. Original link: https://doi.org/10.1016/j.bios.2025.117720 Spotlight 1. Label-free detection for simplified workflow The platform innovatively combines a split aptamer with Auramine O fluorescence, eliminating the need for labeled probes. This reduces cost and minimizes background noise. 2. Dual cascade amplification for enhanced sensitivity By integrating Cas12a-mediated DNAzyme cleavage with aptamer reassembly, the system achieves positive signal amplification, reaching a detection sensitivity of 1.74 nM. 3. Clinical application potential The assay demonstrated effective discrimination in cell lines, CRC patient tissues, and blood samples, showing high concordance with qPCR results. It holds promise for non-invasive early screening of colorectal cancer. Through spectroscopic characterization of the AO/DAP-10 complex, the researchers determined optimal fluorescence parameters (excitation/emission: 480/540 nm). Within the 0-1 μM range, the fluorescence intensity of the complex exhibited a strong linear relationship with DAP-10 concentration (R²= 0.9974), indicating suitability for quantitative detection. When comparing different Cas12a trans-cleavage substrates, DNAzyme showed the highest cleavage efficiency, reaching maximum signal intensity within 15 minutes, significantly faster than linear ssDNA (45 minutes) and G4-structured DAP-10 (90 minutes). The study further confirmed that the split DAP-10 aptamer could effectively reassemble and restore fluorescence in the presence of linker DNA, while the CRISPR/Cas12a system protected the linker by degrading the DNAzyme, thereby activating the fluorescence signal. In methylation detection applications, this system successfully distinguished AciI-treated methylated from unmethylated Septin9 DNA fragments, with methylated samples exhibiting markedly enhanced fluorescence. These results highlight the platform’s high specificity and detection capability. Figure 1. Feasibility analysis of a label-free CRISPR/Cas12a sensor for DNA methylation detection Through systematic optimization, the researchers established the optimal reaction conditions for key components in the detection system: • Auramine O (AO) fluorescence dye: 20 μM achieved maximum signal output. • Linker DNA: 750 nM reached a plateau signal. • DNAzyme: 250 nM provided the best signal-to-noise ratio. • Methylation-specific enzyme AciI: 2 U completely digested unmethylated DNA, minimizing background interference. • Cas12a protein and crRNA: 10 nM each yielded the highest fluorescence signal. Figure 2. Component optimization results Regarding reaction conditions, the study confirmed that linker DNA and split DAP-10 fragments exhibited the highest hybridization efficiency at 20 ℃. To simultaneously ensure enzymatic reaction efficiency, the overall reaction temperature was set to 37 ℃, at which DNAzyme catalytic activity is maximized. Under this temperature, Cas12a trans-cleavage of the DNAzyme was completed within 15 minutes, while DNAzyme-mediated hydrolysis of the linker DNA reached its maximum within 10 minutes. These optimized conditions collectively ensure an optimal balance between sensitivity, speed, and specificity for the detection system. Figure 3. Optimization results of reaction conditions The performance of the Septin9 methylation assay was validated in colorectal cancer cell lines (HCT116 vs. NCM460), paired tissues (tumor vs. adjacent normal), and peripheral blood samples (patients vs. healthy donors). The results showed strong concordance with qPCR measurements (R² = 0.8912), demonstrating the assay’s potential for clinical translation as a non-invasive early screening tool for colorectal cancer. Figure 4. Detection results of clinical samples In this study, the researchers developed a label-free detection platform combining CRISPR/Cas12a, DNAzyme, and split aptamer cascades, successfully enabling sensitive detection of DNA methylation in the key colorectal cancer biomarker Septin9. This approach avoids the use of expensive and complex fluorescently labeled probes and eliminates the damaging effects of conventional bisulfite treatment on DNA, offering a simple and cost-effective workflow. Validation in clinical samples highlights its potential as a non-invasive early screening tool for colorectal cancer. Looking forward, this method could be integrated with microfluidic chips or point-of-care testing (POCT) devices, facilitating deployment in primary healthcare settings and at-home self-testing.









![[Customer Publication] Methylation “Fingerprint” Detection May Transform Early Cancer Screening](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)