[Customer Publication] CRISPR Technology Upgraded! Faster and More Accurate Discovery of Malaria Drug-Resistance Mutations
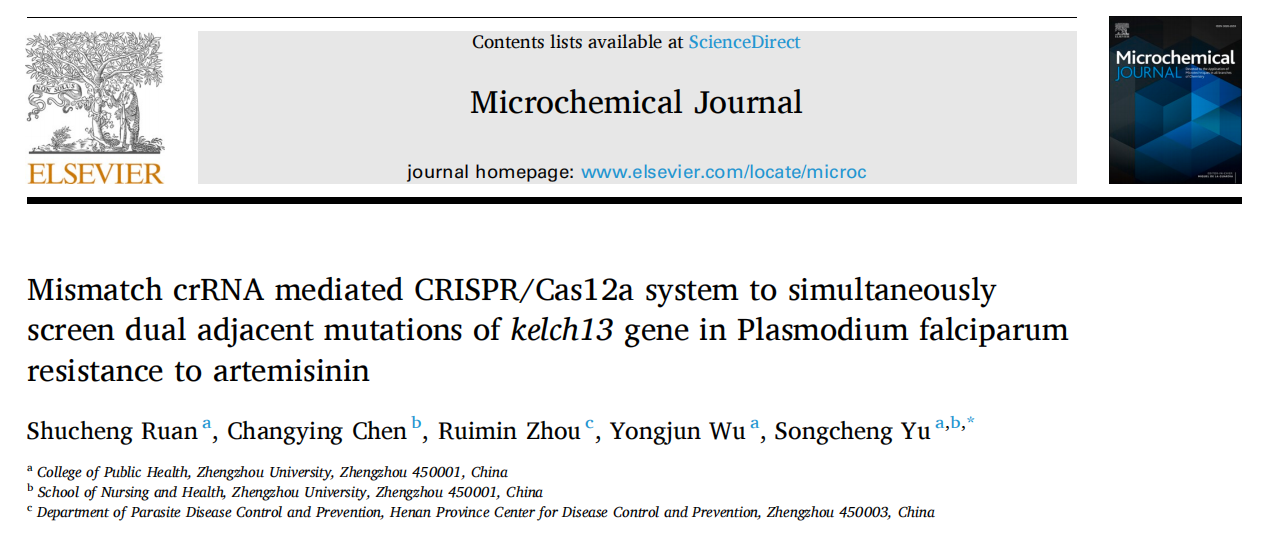
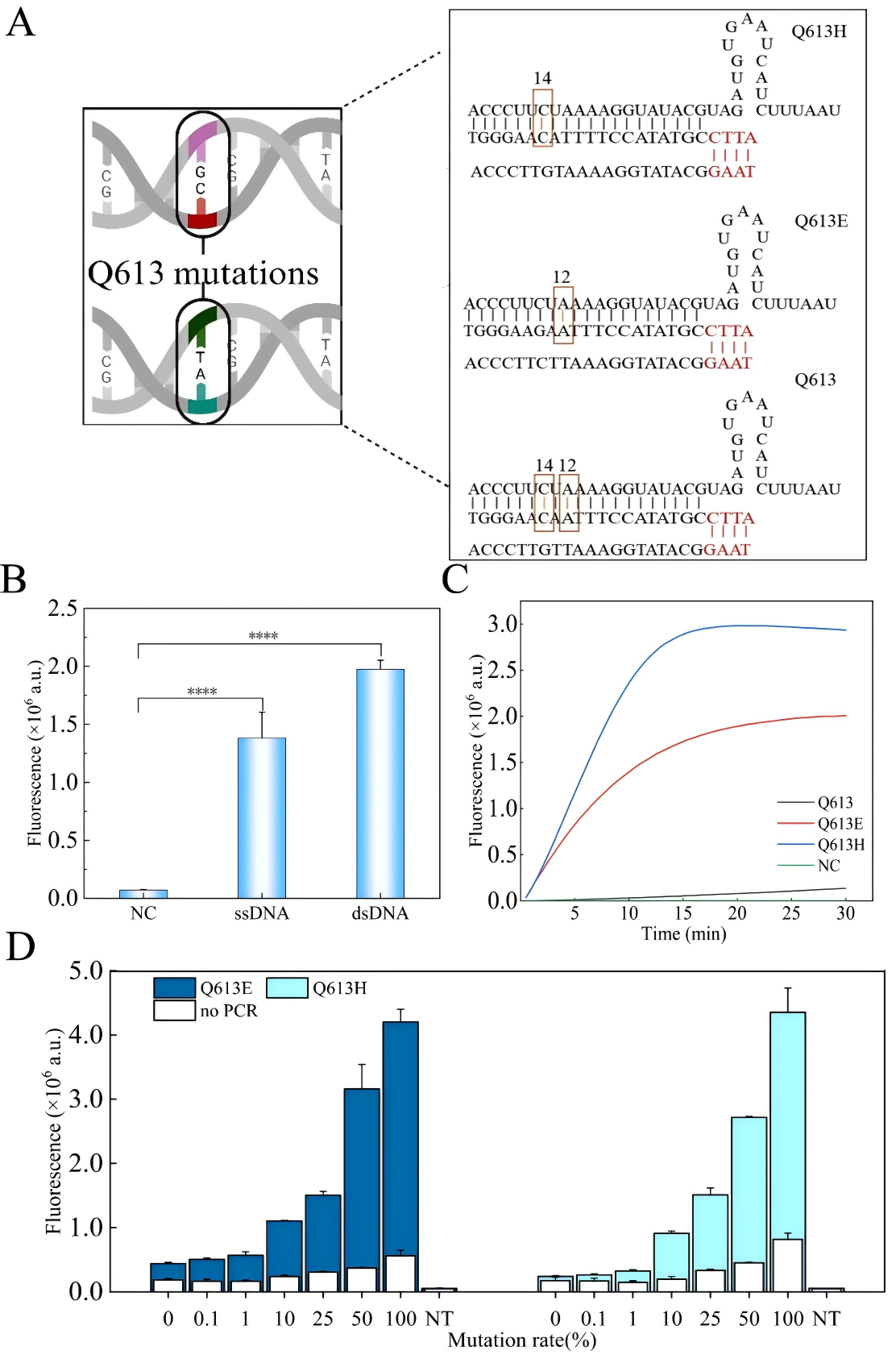
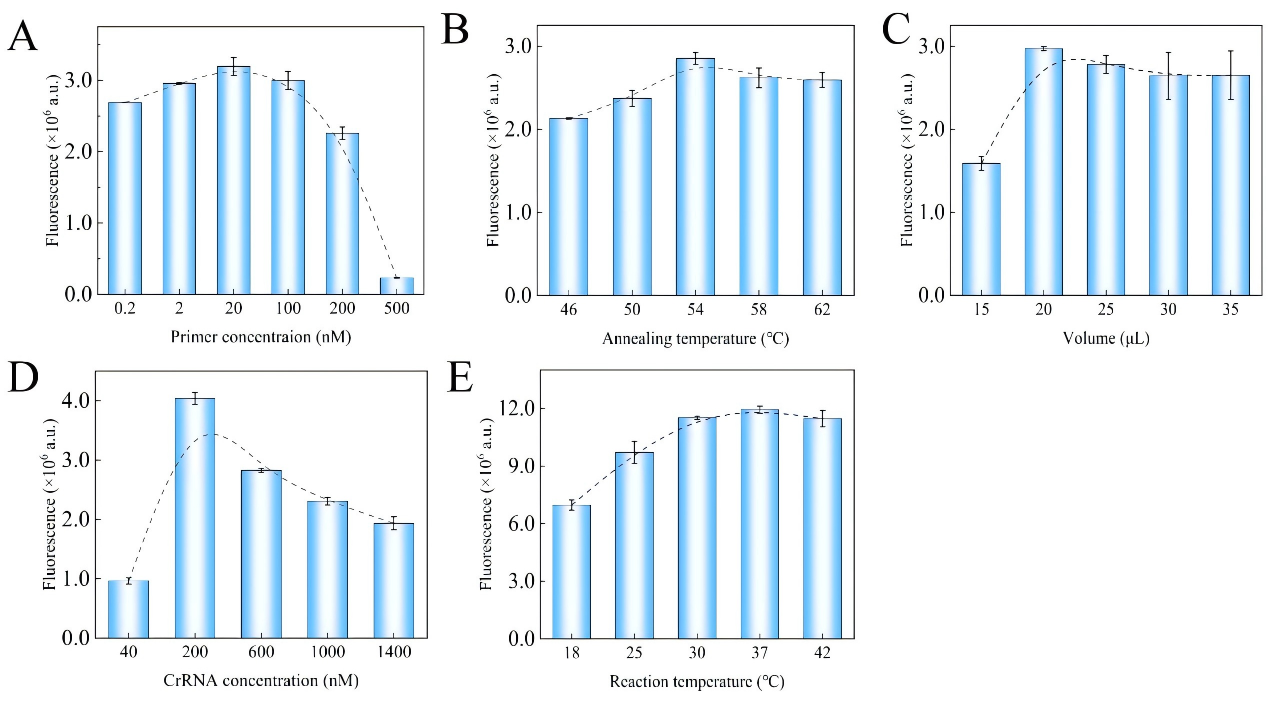
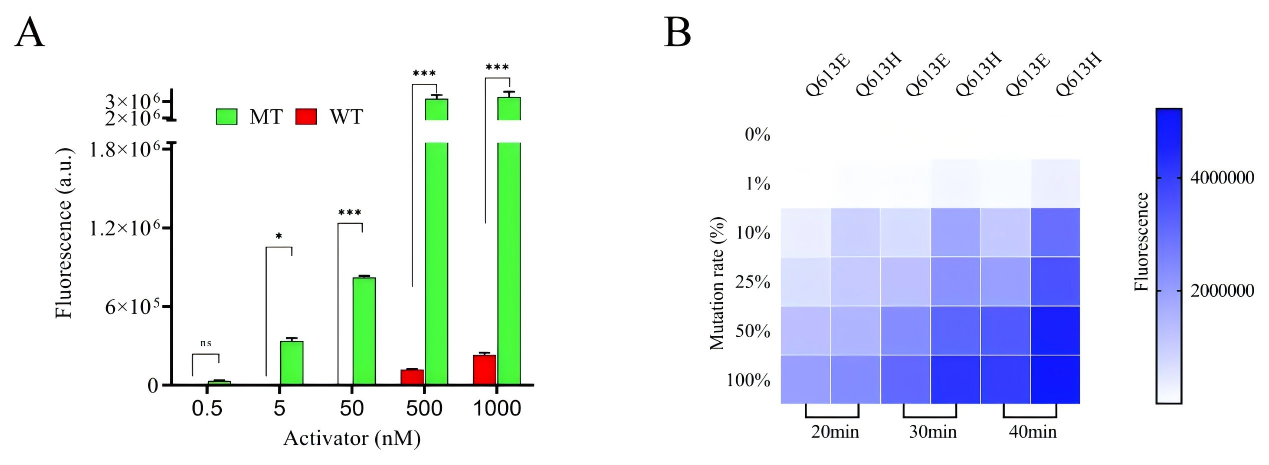
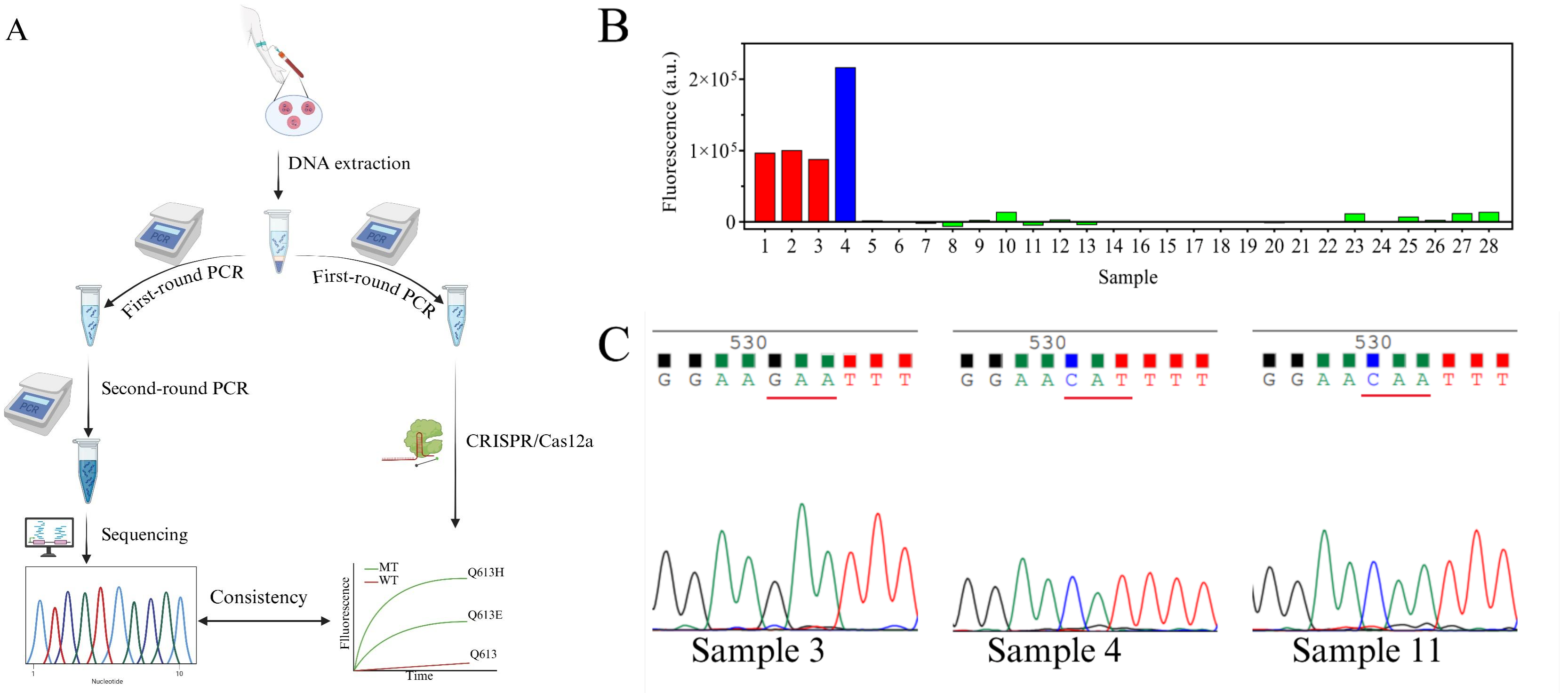
Artemisinin resistance in Plasmodium falciparum is closely linked to mutations in the kelch13 gene, particularly dual-base substitutions at codon 613 (Q613E/Q613H). Traditional PCR and sequencing methods cannot reliably distinguish these mutations at the same site, while existing CRISPR-Cas12a–based multiplex assays often rely on additional equipment such as quantum dots or microfluidic chips, making them difficult to deploy in resource-limited settings. A research team led by Chinese scientists, recently published a paper in Microchemical Journal entitled “Mismatch crRNA mediated CRISPR/Cas12a system to simultaneously screen dual adjacent mutations of kelch13 gene in Plasmodium falciparum resistance to artemisinin.” In this work, the team developed a novel “mismatch crRNA” strategy that enables one-pot discrimination of Q613 wild-type, Q613E, and Q613H variants. This approach allows rapid, low-cost, and highly sensitive genotyping of single amino acid substitutions at codon 613 of kelch13, providing a powerful tool for monitoring artemisinin resistance in malaria-endemic regions such as Africa, and offering a paradigm for detecting complex mutations in other pathogens. EDITGENE is proud to have supported this research by providing high-performance Cas12a proteins, contributing to the successful outcome of the study. Original link: https://doi.org/10.1016/j.microc.2025.114209 Spotlight 1. Technical innovation: mismatch crRNA + CRISPR/Cas12a The study introduces a mismatch crRNA strategy that enables the CRISPR/Cas12a system to distinguish three kelch13 genotypes—Q613, Q613E, and Q613H—within a single-tube reaction. 2. Detection advantages: sensitive, rapid, and cost-effective The assay delivers results in just 30 minutes, detects mutations at concentrations as low as 20 pM, and accurately differentiates dual mutations with outcomes consistent with Sanger sequencing, making it practical for use in resource-limited settings. 3. Application potential: malaria resistance surveillance and control Validated in 28 clinical samples, the method provides an effective tool for monitoring artemisinin resistance–associated mutations and offers scalability to other drug-resistance loci. Experimental validation confirmed that mismatch-designed crRNAs can specifically distinguish between wild-type and mutant kelch13 Q613 variants (Q613E/Q613H). For the wild-type Q613, the presence of double mismatches failed to activate Cas12a, yielding signals comparable to the negative control. In contrast, Q613E and Q613H produced strong fluorescence signals, with Q613H generating a higher signal intensity due to its mismatch position being farther from the PAM site. For low-abundance mutations, PCR pre-amplification was required for effective discrimination. The results demonstrated high specificity and improved detection sensitivity, enabling reliable identification of mutations with frequencies as low as 0.1%. In summary, the mismatch crRNA-mediated CRISPR/Cas12a system allows highly specific detection of Q613 mutations, though trace-level detection remains dependent on pre-amplification. Figure 1. Feasibility results of mismatch crRNA–mediated CRISPR/Cas12a Through systematic optimization, the researchers established a high-performance CRISPR/Cas12a detection system. Key parameters include: • A PCR primer concentration of 20 nM yielded the strongest signal, while excess primers competitively suppressed Cas12a trans-cleavage activity. • An annealing temperature of 54 °C provided maximal amplification efficiency. • A PCR reaction volume of 20 μL was identified as optimal. • A crRNA concentration of 200 nM produced the highest fluorescence signal. • A reaction temperature of 37 °C maintained Cas12a activity while preserving mismatch-specific recognition. Together, these optimized conditions significantly enhanced the overall performance and reliability of the detection system. Figure 2. Optimization results The mismatch crRNA-mediated CRISPR/Cas12a system demonstrated a detection limit of 20 pM for the Q613E mutant, with clear discrimination from the wild-type Q613. In mixed samples, the assay successfully identified Q613E and Q613H mutations at frequencies as low as 1%, with Q613H generating a stronger fluorescence signal due to the mismatch distance effect. Notably, this method achieved simultaneous differentiation of two adjacent mutations (Q613E and Q613H) within a single-tube assay, without reliance on complex instrumentation. These findings highlight its potential as a point-of-care testing (POCT) tool. Figure 3. Performance evaluation of the method The mismatch crRNA-mediated CRISPR/Cas12a system was applied to genotype the kelch13 gene in 28 clinical samples: • Pre-amplification followed by gel electrophoresis confirmed successful amplification of the target fragment in all samples. • CRISPR-based detection revealed strong mutation signals in samples 1-4. Among these, samples 1-3 showed comparable fluorescence intensities, consistent with the Q613E mutation, while sample 4 exhibited a markedly higher signal intensity, indicative of the Q613H mutation. Samples 5-28 were identified as wild-type Q613. • Validation by Sanger sequencing showed 100% concordance with the CRISPR assay results. These findings demonstrate the excellent accuracy and reliability of the proposed method in real clinical samples, underscoring its potential as a robust tool for on-site surveillance of artemisinin resistance in malaria-endemic regions. Figure 4. Detection results of clinical samples In this study, the researchers established a CRISPR/Cas12a-based detection platform leveraging mismatch-designed crRNAs, enabling one-pot and rapid discrimination of the kelch13 gene Q613, Q613E, and Q613H variants. The method combines high sensitivity, rapid turnaround, and low cost, offering a practical solution for resistance surveillance in resource-limited regions. Looking forward, this strategy could be extended to other key loci associated with antimalarial drug resistance. Coupled with field-deployable devices and portable readout systems, the platform has the potential to evolve into a ready-to-use tool for frontline healthcare and malaria control. Ultimately, it may provide critical support for early warning of resistance and contribute to global strategies for malaria elimination.