[Literature Review] Cas9 Safety Breakthrough: Reducing Off-Targets from 77% to Zero Translocations
Gene Editing

Chronic granulomatous disease (CGD) is caused by defects in NADPH oxidase. Current stem cell transplantation therapies carry significant risks, including graft-versus-host disease, graft failure, and severe infections. Although CRISPR/Cas9-mediated HDR gene therapy offers the potential for autologous transplantation, it still faces major safety concerns such as off-target effects, chromosomal abnormalities, and p53 activation.
Recently, Jacob Giehm Mikkelsen and Rasmus O. Bak’s teams at Aarhus University (Denmark) published a study in Nature Communications titled “Targeted gene editing and near-universal cDNA insertion of CYBA and CYBB as a treatment for chronic granulomatous disease”.
In this work, the researchers employed a high-fidelity Cas9 variant combined with paired Cas9 nickases to efficiently repair the CYBB and CYBA genes in CD34⁺ cells from CGD patients. This approach markedly reduced off-target effects and cytotoxicity, while eliminating chromosomal translocation events. Importantly, the gene edited cells demonstrated long-term multilineage engraftment and restored granulocyte function in vivo, providing a safer and more effective therapeutic strategy with strong potential for clinical translation.

Original link: https:// doi.org/ 10.1038/s41467-025-62738-2
Spotlight
1. Multipronged Optimization of HDR
For the first time in a CGD model, the authors co-delivered mRNAs encoding i53 (53BP1 inhibitor), GSE56 (p53 inhibitor), and Ad5-E4orf6/7 (HDR enhancer). This combinatorial strategy significantly boosted editing efficiency and preserved cell viability within long-term hematopoietic stem cell (LT-HSC)-enriched populations.
2.Toward a “Pan-X-CGD” Therapeutic Strategy
The researchers designed a targeted insertion of CYBB exon 3-13 cDNA, a strategy theoretically capable of correcting ~86% of X-CGD mutations. This approach provides a feasible pathway for developing a universal gene therapy for X-CGD.
3. Stepwise Safety Upgrades
By moving from Std-Cas9 to HiFi Cas9 and ultimately to paired D10A Cas9 nickases, the researchers progressively reduced off-target editing from ~77% to undetectable levels. Using CAST-seq analysis, they further confirmed that the paired nickase strategy effectively eliminated chromosomal translocations.
The researchers first tested a standard workflow—delivering Std-Cas9 RNPs together with an rAAV6 repair template—to target CYBA with a GFP reporter insertion. While this approach achieved relatively high editing efficiency (~40%), it triggered a strong DNA damage response in hematopoietic stem and progenitor cells (HSPCs), leading to reduced proliferation and a marked decline in colony-forming capacity (CFU).
Most critically, when these edited cells were transplanted into immunodeficient mice, they almost completely lost long-term, multilineage engraftment potential. Edited cell frequencies in recipient bone marrow dropped sharply, indicating that the editing process imposed strong negative selection or direct damage on bona fide LT-HSCs.
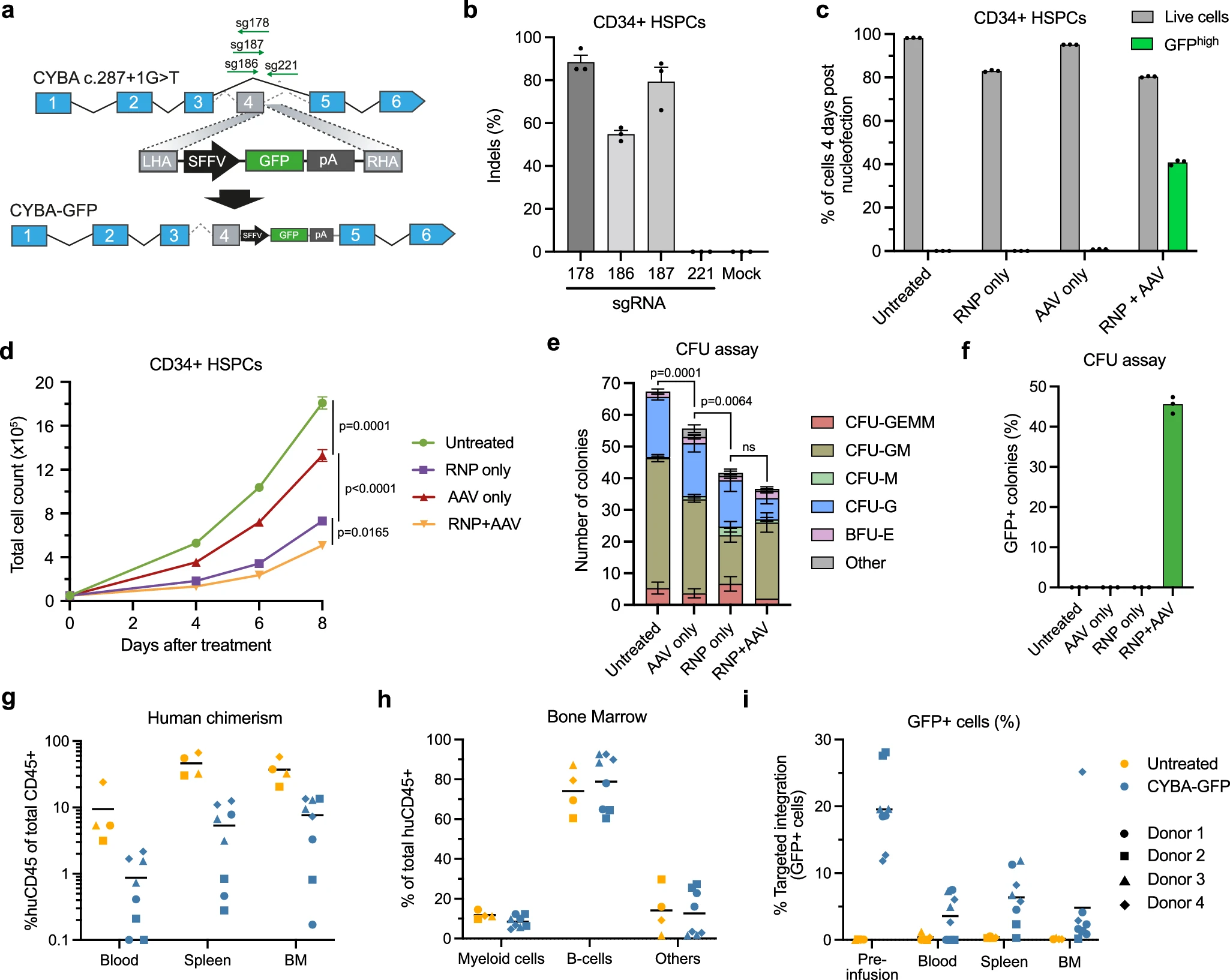
Figure 1. Targeted CYBA insertion using RNP + AAV
To overcome the observed toxicity, the researchers supplemented the editing system with three mRNAs: i53, GSE56, and Ad5-E4orf6/7. Co-delivery of these HDR enhancers markedly increased HDR efficiency in LT-HSC-enriched populations, while also boosting the overall number and viability of edited cells. This strategy not only improved repair efficiency but also alleviated the detrimental impact of genome editing on LT-HSC fitness.
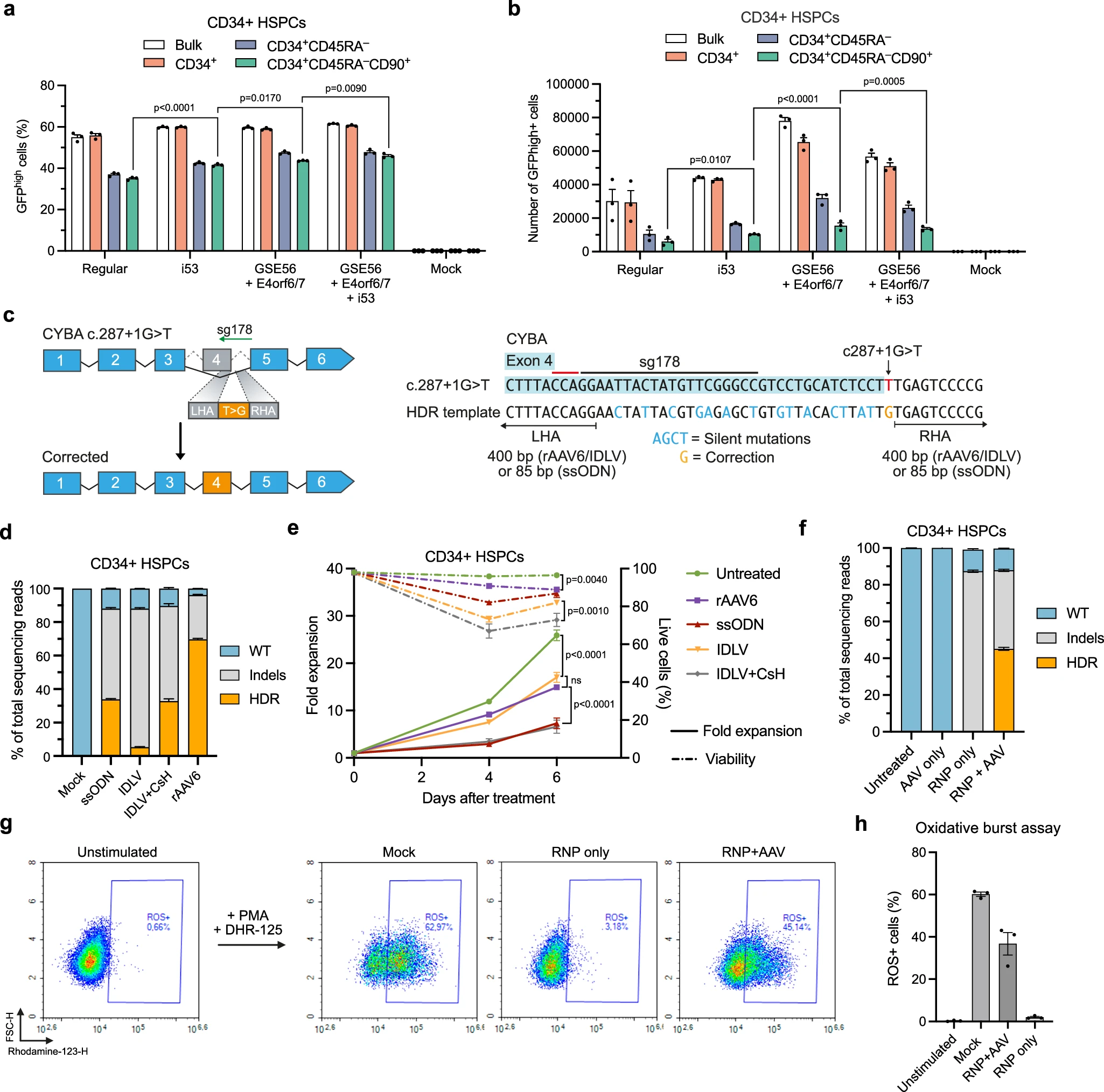
Figure 2. Optimized CYBA gene editing strategy
Focusing on the CYBB c.252G>A mutation, the researchers observed that using a non-specific sgRNA combined with a repair template carrying silent mutations yielded higher correction efficiency. Building on this, they developed a universal approach: HDR-mediated insertion of a truncated cDNA fragment (exons 3-13), theoretically correcting the majority of X-CGD mutations. Edited cells successfully restored ROS production under this strategy.
Initially, the sg3/Std-Cas9 setup led to severe off-target activity (OT1 up to 77.5%) and extremely poor engraftment in vivo (<1%). Switching to HiFi Cas9 reduced off-target rates to below 5% while maintaining on-target efficiency. Edited cells also showed markedly improved CFU capacity in vitro, and in animal models achieved robust long-term, multilineage engraftment at levels up to ~67%.
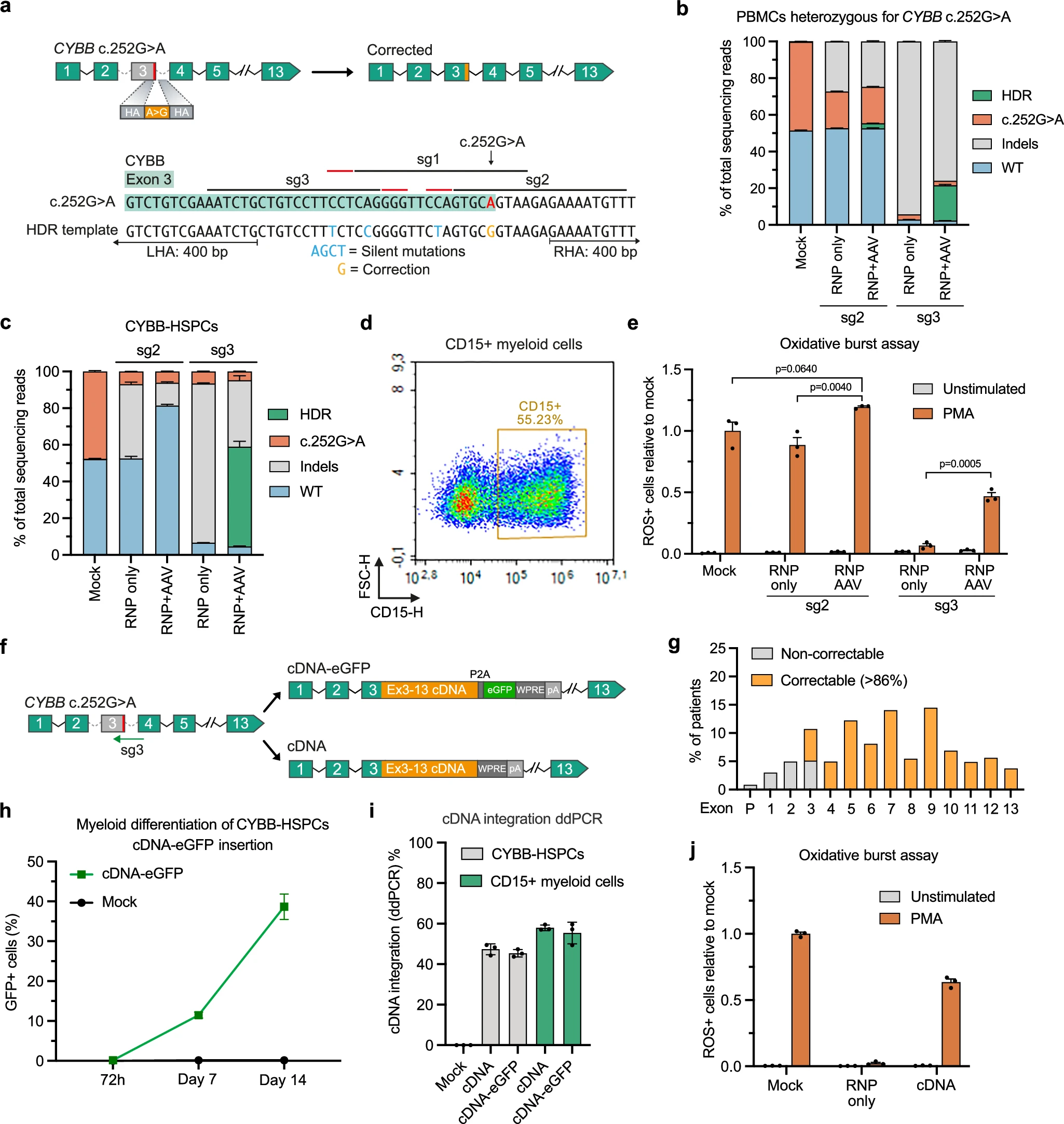
Figure 3. Allele-specific editing and cDNA insertion for CYBB correction
To further enhance safety, the researchers employed a paired D10A Cas9 nickase strategy, using two sgRNAs to create adjacent single-strand cuts that mimic a double-strand break. This approach markedly reduced off-target activity compared with standard nucleases.
In animal models, the optimized nickase strategy proved effective: edited cells achieved durable engraftment while maintaining stable editing levels over time. These results demonstrate that the paired nickase system can efficiently and safely edit LT-HSCs, supporting long-term and stable expression of therapeutic transgenes.
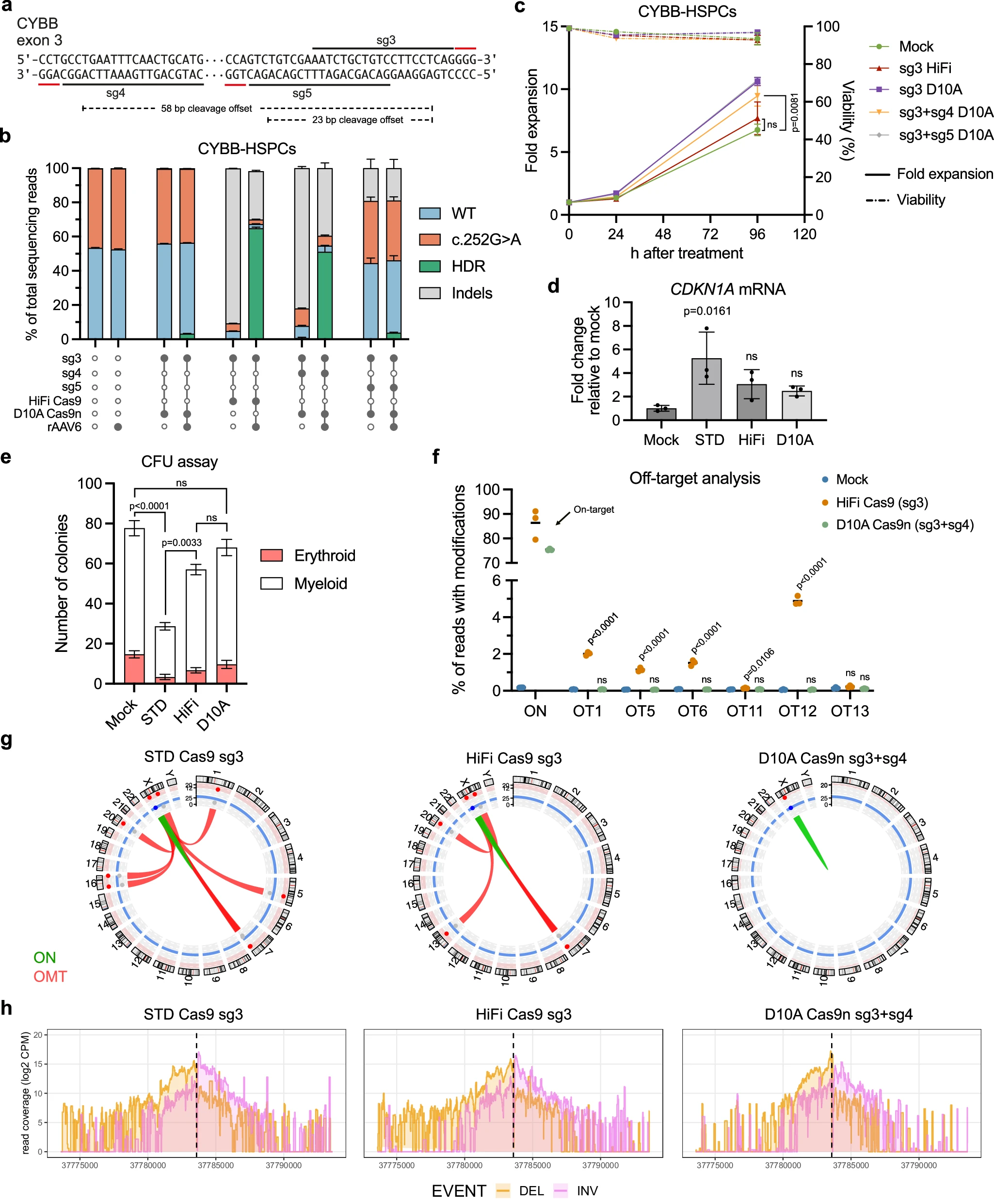
Figure 4. Safety and targeting efficiency of paired D10A Cas9 nickase editing
This study demonstrates that combining high-fidelity Cas9 variants with paired Cas9 nickase systems enables efficient and safe editing of genes associated with chronic granulomatous disease (CGD). Edited cells exhibited durable engraftment and stable long-term correction in animal models, offering a promising new avenue for CGD gene therapy.
Looking ahead, further improvements are needed to enhance HDR efficiency within LT-HSCs and to minimize editing-related toxicity, paving the way for safe and scalable clinical applications.

















