[Literature Review] CRISPR screening system reveals synthetic lethality in DNA damage response
CRISPR Screening
CRISPRi

Comprehensive CRISPR Screening Reveals Vulnerabilities in the DNA Repair Network


Original link:https://doi.org/10.1038/s41586-025-08815-4

To fully explore the redundancy and compensatory mechanisms within the DNA damage response (DDR) pathway, researchers developed a dual-guide CRISPRi library named SPIDR (Systematic Profiling of Interactions in DNA Repair). The library targets 548 core DDR genes, generating nearly 700,000 sgRNA combinations, and was screened in RPE-1 cells. Using CRISPR interference instead of Cas9 cutting, the system avoids introducing DNA breaks, making it ideal for studying gene pairs essential to cell survival.
By applying the GEMINI variational Bayesian algorithm to model screening data at scale, the researchers identified around 5,000 synthetic lethal interactions (GEMINI score ≤ –1). These interactions span key biological processes such as DNA replication, repair, chromatin remodeling, and transcriptional regulation. The SPIDR library not only confirmed known interactions like BRCA2 and LIG1, but also uncovered many novel functional links. This platform offers a powerful tool for building genetic interaction maps under homeostatic conditions.
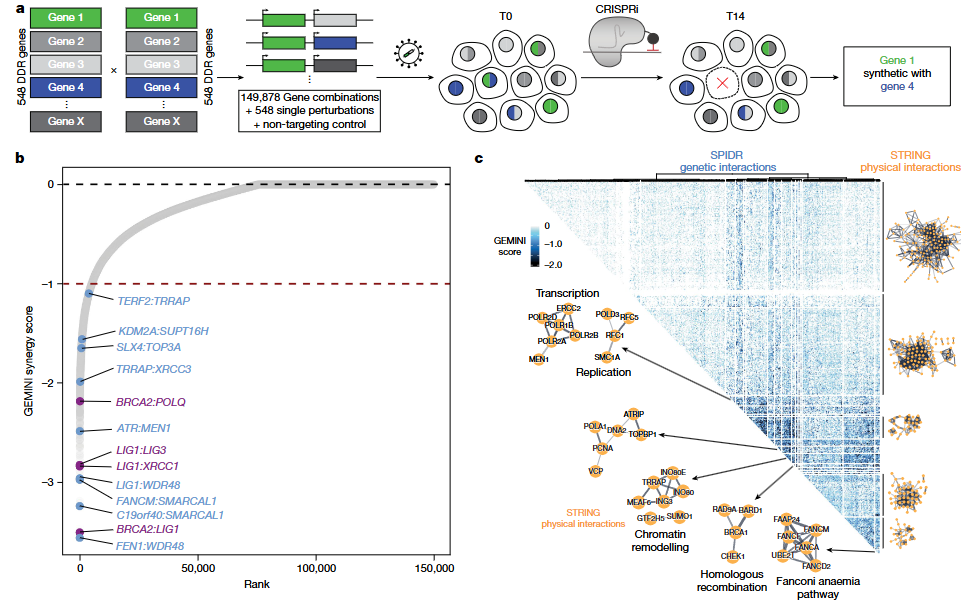
Figure 1. CRISPR interference-based screening of 548 core DNA repair genes

Among the identified interactions, the researchers focused on two high-scoring synthetic lethal gene pairs that highlight distinct roles in preserving genome integrity.
First, the synthetic lethality between FEN1/LIG1 and WDR48–USP1 stems from disrupted PCNA ubiquitination.
When FEN1 or LIG1 is lost, unsealed gaps accumulate during DNA replication and require alternative repair pathways. The WDR48–USP1 complex normally removes polyubiquitin chains added by RAD18 to PCNA, maintaining its stability. Without this regulation, PCNA is degraded, leading to replication stress, chromosomal breaks, and cell death. Notably, cells with compromised FEN1 or LIG1 function showed strong sensitivity to USP1 inhibitors, suggesting possible therapeutic value.
Second, FANCM and SMARCAL1 are DNA translocases that help remove cruciform-like DNA structures in AT-rich regions. Dual loss prevents proper resolution of these structures, which are then misrecognized and cleaved by the ERCC1–ERCC4 endonuclease, resulting in chromosomal breaks. This process does not rely on replication fork reversal, pointing to a novel source of structural genome instability.
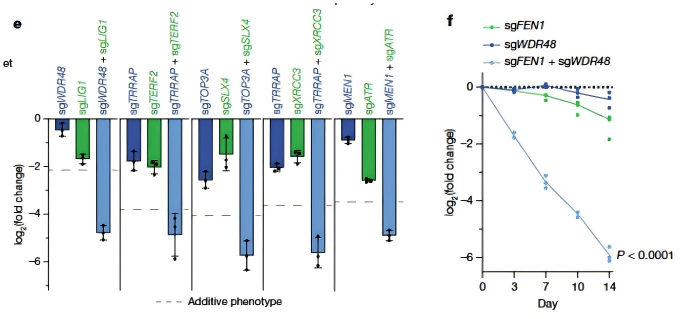
Figure 2. Dual-sgRNA competitive growth assays confirm the synthetic lethality of LIG1:WDR48 and FEN1:WDR48 pairs
From Screening to Therapy: Synthetic Lethality Networks Drive Precision Oncology

Another key contribution of this study lies in integrating the synthetic lethality interaction map with cancer mutation data (COSMIC) and drug target databases (DGIdb) to identify gene–compound pairs with therapeutic potential. For example, bladder cancer cells with ERCC2 mutations may be sensitive to DNA-PKcs inhibitors, while FEN1-mutant tumors could show increased reliance on USP1 inhibition. In addition, mutations in FANCM are found across various cancer types, pointing to SMARCAL1 as a possible therapeutic target.
As CRISPR technologies continue to advance, systematic functional screening platforms like SPIDR are becoming essential tools for uncovering disease vulnerabilities. Synthetic lethality maps offer a quantifiable and testable molecular framework—particularly valuable for drug combination design, resistance mechanism analysis, and personalized treatment strategies.
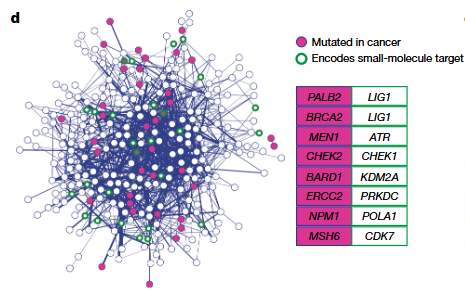
Figure 3. Synthetic lethality network map highlighting gene pairs that overlap with cancer mutations and small-molecule drug targets
This study highlights the powerful potential of dual-guide CRISPRi interference screening combined with high-throughput analytics to dissect complex genetic networks. By uncovering synthetic lethal interactions under baseline conditions, the SPIDR platform addresses key blind spots left by conventional CRISPR knockout screens and proposes several actionable strategies for cancer therapy. Looking ahead, similar technologies are poised to expand their impact across oncology, neurodegenerative diseases, and immune system disorders, paving the way for more refined and personalized genomic interventions.
















