Alextina | Dec 28,2024
Pre-experiment essentials: A walkthrough of the CRISPR Assay
Principle of CRISPR detection
CRISPR detection technology is a means of detection developed based on the principle of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology. It is a process in which the target nucleic acid activates the trans-cutting activity of Cas protein, shredding the fluorescently labeled probe in the system and emitting a fluorescent signal to complete the detection of the target nucleic acid. This technology utilizes the specific recognition and unique subsidiary cutting ability of the CRISPR-Cas system to achieve efficient, rapid, and sensitive detection of target DNA or RNA sequences. CRISPR detection has several significant advantages, including high sensitivity, high specificity, rapid editing, economical and practical, etc. Therefore, it has been widely used in various fields, such as environmental testing, animal epidemiology, disease monitoring, pet diseases, food safety, and so on. CRISPR is widely used in many fields, such as environmental testing, animal disease, disease monitoring, pet disease, food safety, etc.
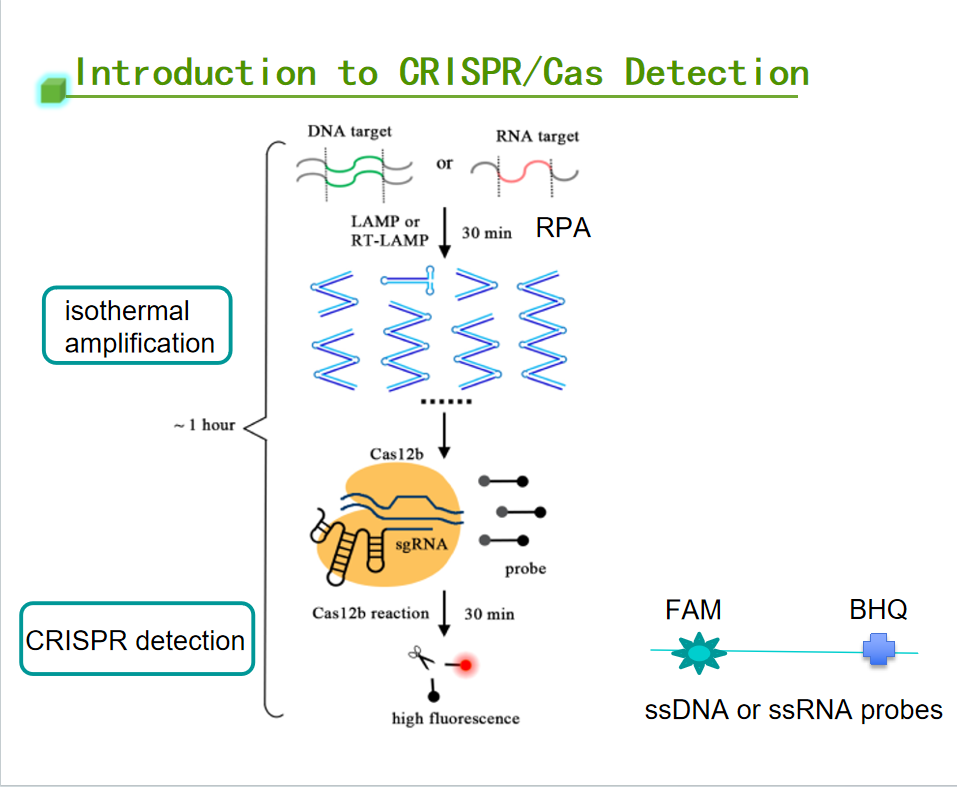
EDITGENE has compiled the crucial knowledge points of CRISPR detection below. May it contribute to and expedite your research.
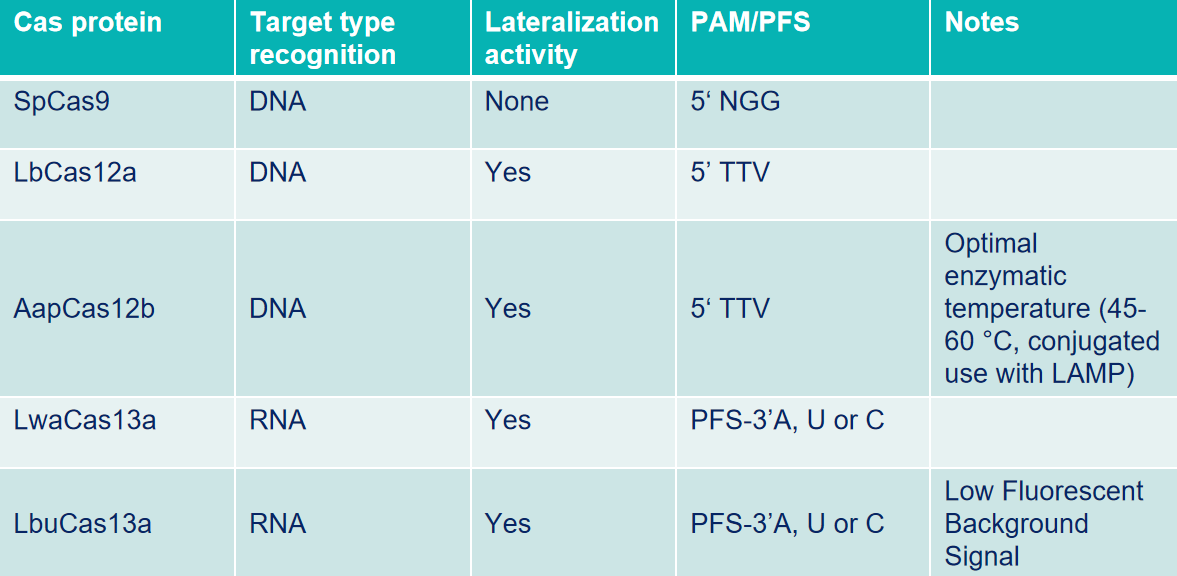
Selection of Cas enzyme
Cas enzyme plays a crucial role in CRISPR detection. It can accurately identify and cut DNA or RNA that is complementary to the target sequence, thereby achieving efficient detection of specific genes. Due to differences in experimental purposes, target genes, operating conditions, etc., selecting the appropriate Cas enzyme is also particularly important.
Cas12a: Also known as Cpf1, is an endonuclease that can specifically recognize and cut double-stranded DNA (dsDNA) targets with PAM (5'-TTTN-3' or 5'-TTN-3'). Breaks DNA double strands and generates sticky ends. Furthermore, recognition and cleavage of single-stranded DNA (ssDNA) targets are independent of PAM sequences.
Cas12b: Also known as a sgRNA-mediated endonuclease that can specifically recognize and cut dsDNA with PAM (5'-TTN-3), causing DNA double-strand breaks and generating sticky ends. In particular, it has higher cutting activity in medium and high-temperature environments of 45-65°C.
Cas13a: Guided by sgRNA, Cas13a can recognize and cleave specific single-stranded RNA in the system. After cutting a specific RNA sequence, its "accessory cleavage" activity will be activated, efficiently cutting any ssRNA sequence in the system.
- EDITGENE has independently developed a protein expression purification platform that has obtained high purity, high activity, and high recovery Cas12/Cas13 enzymes , with detection limits up to the amol level!
crRNA design and synthesis:
crRNA is a key component of the CRISPR-Cas system. It recognizes and guides the Cas protein to the target nucleic acid sequence position. It also guides the Cas protein to perform precise cutting or editing through complementary pairing with the target DNA or RNA sequence. crRNA greatly affects the sensitivity of the detection system, so we need to choose the crRNA with the best effect in the experiment. The design and synthesis of crRNA involves the following precautions:
1. Target sequence selection: Select a specific target sequence as the starting point for design, usually determined based on the function of the target gene and the needs of related research, and select high-copy repetitive segments in the genome as the target gene.
2. PAM sequence selection: Select an appropriate PAM sequence near the target sequence.
3. Avoid cutting sites: Avoid cutting sites that match other non-target gene sequences. This can be accomplished by using sequence alignment software.
4. Avoid secondary structure at the cutting site: avoid the target sequence and other non-target sequences from forming a stable secondary structure
5. Sequence selection and verification: During the design process, sequence comparison tools and structure prediction tools can be used to evaluate the specificity and effectiveness of the designed crRNA. Finally, the specificity and cutting efficiency of the designed crRNA are confirmed through experiments.
The following are our commonly used crRNA primer design sequences:
1. lbCas12a:
5'AAUUUCUACUAAGUGUAGAUNNNNNNNNNNN 3' (N denotes the spacer region that is complementarily paired with the specific target sequence)
2.AapCas12b:
5'-GUCUAGAGGACAGAAUUUUUCAACGGGUGUGCCAAUGGCCACUUUCCAGGUGGCAAAGCCCGUUGAG
CUUCUCAAAUCUGAGAAGUGGCACNNNNNNNNNNNNNNN-3' (N denotes the spacer region that is complementarily paired with the specific target sequence)
3.LwaCas13a:
5'GAUUUAGACUACCCCAAAAACGAAGGGGACUAAAACNNNNNNNNNNNNNNNNNNN3' (N denotes spacer region complementarily paired with the specific target sequence, recommended spacer region length is 20-28 nt)
4.LbuCas13a:
5'GGACCACCCCAAAAAUGAAGGGGACCAAAACNNNNNNNNNNNNNNNNNNNN-3’(N denotes the spacer region that is complementary to the specific target sequence)
Selection of Isothermal Amplification System
Isothermal Amplification is one of the key methods for rapid and accurate detection. It is easier and faster than the traditional PCR method, especially in the need for rapid on-site detection or limited equipment conditions. The following is an introduction to common thermostatic amplification systems.
1. LAMP (Loop-mediated isothermal amplification)
Characteristics: The LAMP technique relies on a set of tailored primers and the specific cleavage activity of Bst DNA polymerase, which can be performed at a constant temperature (usually 60°C-65°C). This method allows for efficient amplification in a short period of time (typically less than an hour). It allows visual observation of the amplification results (e.g., the adding fluorescent agent).
2. RPA (Recombinase Polymerase Amplification)
Characteristics: RPA technology works at or near room temperature (37°C-42°C) and requires no special equipment. PA uses recombinant enzymes to unravel double-stranded DNA and amplify the target DNA in a short period of time.
3. NASBA (Nucleic Acid Sequence Dependent Amplification)
Characteristics: NASBA reaction is carried out at constant temperature (41°C-42°C), and the amplified product can be single-stranded DNA or double-stranded RNA. A lower concentration of RNA targets can be detected, but the stability of reagents is required to be higher, and the operation is relatively complicated.
When selecting a thermostatic amplification system, a variety of factors need to be considered, including the purpose of the experiment, the characteristics of the target sequence, the experimental conditions, and the stability and efficiency of the amplification system. The following are two important factors to be considered when selecting a thermostatic amplification system:
(1)Characteristics of the target sequence: The length of the target sequence, GC content, the existence of secondary structure, and other factors will affect the effect of thermostatic amplification. Therefore, when choosing a thermostatic amplification system, it is necessary to fully understand the characteristics of the target sequence and choose an amplification system suitable for those characteristics.
(2) Stability and efficiency of the amplification system: The stability and efficiency of the thermostatic amplification system are important factors to be considered in the selection. A stable system can reduce experimental errors and improve the accuracy of the results. An efficient system can shorten the experimental time and improve the experimental efficiency.
As a "next-generation molecular diagnostic technology," the CRISPR assay significantly impacts the specificity and sensitivity of subsequent experiments by choosing the right Cas enzyme, optimal crRNA sequence, and constant-temperature amplification system before conducting the experiments.
- EDITGENE provides a one-stop supply of high-performance CRISPR assay products, including cas12/cas13, crRNA design and synthesis , thermostable amplification reagents, etc. These can be used in CRISPR assay R&D, genetic testing, animal disease testing, human disease testing, and food safety testing.
Recent News:
[Frontier Information] The first AI gene editor has successfully edited the human genome
[Star of the Month]Human Epigenetic Knockout Library, Mouse & Human Whole Genome Knockout/ Activation Library
[Literature Review] Improving the Sensitivity of LbuCas13a Detection using Engineered crRNAs
Contact us:
+1(224) 345-1927 (USA)
+86-13250544625 (Intl)


 Login
Login









Comment (4)