[Cutting-Edge News] Developments in Prime Editing - Prime Editing Offers New Approaches for Disease Treatment
prime editing

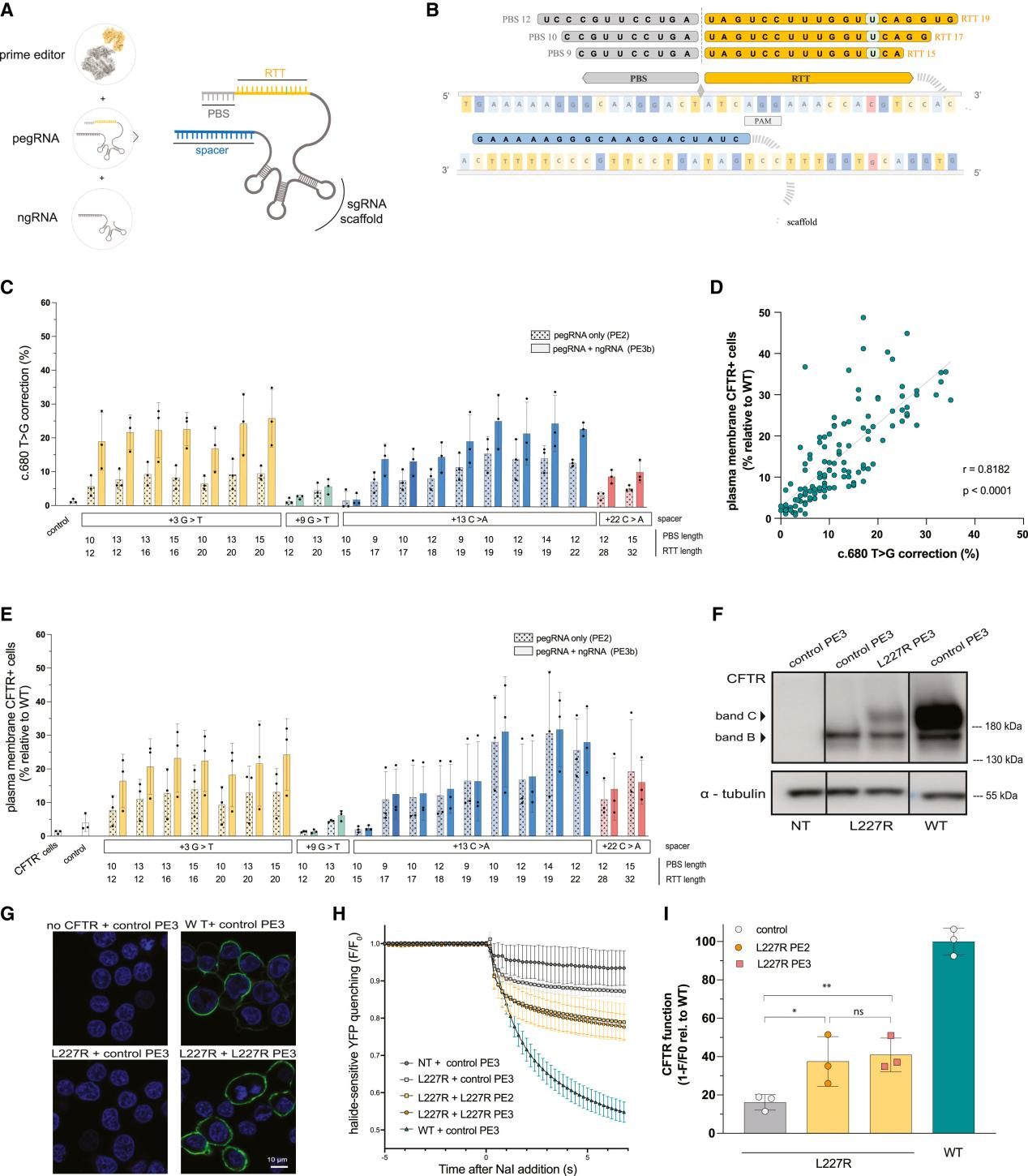
Figure 1 Prime Editing for c.680T>G (L227R) Correction
II. CRISPR Prime Editing Assists in Generating Human Induced Pluripotent Stem Cell Lines
Original Article Link : https://doi.org/10.1089/crispr.2023.0066
With the advancement of human molecular genetics and genomics, thousands of gene loci associated with common disease risks have been identified, often containing multiple candidate variants. However, most disease-related variants are non-coding, making it challenging to identify the molecular mechanisms behind these variants.
The development of CRISPR technology has provided more precise methods for targeted genome editing, particularly prime editing, which can mediate almost any single nucleotide substitution.
Consequently, many researchers aim to utilize prime editing to study the functional relevance of specific gene variants and generate isogenic series in pluripotent stem cells to control genetic background while assessing the dosage effects of causal alleles.
In this article, researchers developed an efficient CRISPR prime editing protocol for generating cell lines carrying heterozygous or homozygous alleles in induced pluripotent stem cells (iPSCs) and optimized the prime editing technique.
They then selected six single nucleotide variants (SNVs) associated with type 2 diabetes (T2D) risk for editing. Editing was performed on iPSCs derived from human donors with different genetic backgrounds for each locus, evaluating editing efficiency through a series of experiments and comparisons.
The study results showed that researchers successfully generated 27 edited iPSC clones covering six SNVs associated with type 2 diabetes or congenital hyperinsulinism (CHI). They also found that prime editing efficiency in iPSCs was higher than in HEK293T cells.
Overall, this study demonstrated the potential of prime editing technology in generating iPSCs with specific genetic backgrounds and provided a powerful tool for studying the impact of specific genetic variants on disease.
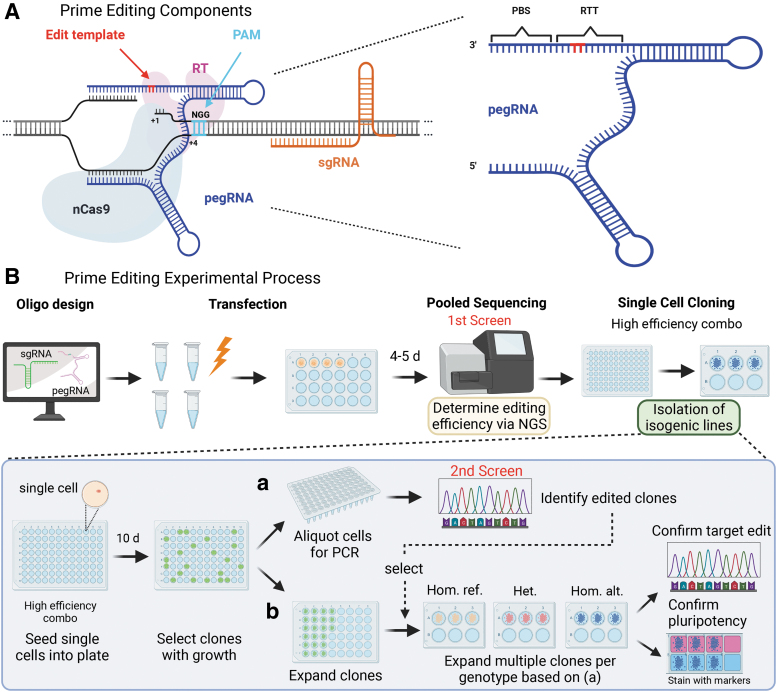
Figure 2 Prime Editing (PE3) Components and Experimental Process
III. In Vitro Prime Editing of Patient-Derived Hematopoietic Stem Cells Rescues Sickle Cell Disease Phenotype in Transplanted Mice
Original Article Link : https://doi.org/10.1038/s41551-023-01026-0
Sickle Cell Disease (SCD) is an autosomal recessive genetic disorder caused by a point mutation from A·T to T·A in the β-globin gene (HBB). Currently, the only FDA-approved cure for SCD is allogeneic hematopoietic stem cell transplantation. However, most patients lack suitable donors, and the procedure can lead to severe toxicity.
By correcting the patient's own hematopoietic stem cells (HSCs), one can bypass immune complications and eliminate the need for tissue-matched donors. Clinical trials are underway using Cas9 nuclease-mediated homology-directed repair (HDR) and adeno-associated virus type 6 (AAV6) delivery of DNA templates to correct SCD mutations.
In this study, researchers used an optimized prime editing system to correct SCD patient-derived hematopoietic stem and progenitor cells (HSPCs) in vitro. They employed electroporation to combine PEmax mRNA with synthesized epegRNA and cutting sgRNA, successfully correcting the SCD allele (HBBS) back to the wild-type (HBBA), with correction frequencies ranging from 15% to 41%.
The edited HSPCs were then transplanted into immunodeficient mice. After 7 weeks, the edited HSPCs maintained HBBA levels in the mice's bone marrow and exhibited similar engraftment frequency, hematopoietic differentiation, and lineage maturation as unedited healthy donor HSPCs.
Evaluation of therapeutic effects showed that, in the transplanted mice, an average of 42% of erythroid progenitors and reticulocytes expressed HBBA, surpassing the predicted therapeutic benefit level. Gene-specific analysis also indicated high target DNA specificity of the prime editing system.
The final results confirmed the safety and efficiency of this technique, demonstrating long-term effects and multi-clonality.
Overall, this study highlights the potential of prime editing technology in treating SCD, proving its effectiveness in enhancing therapeutic outcomes and reducing off-target editing risks.
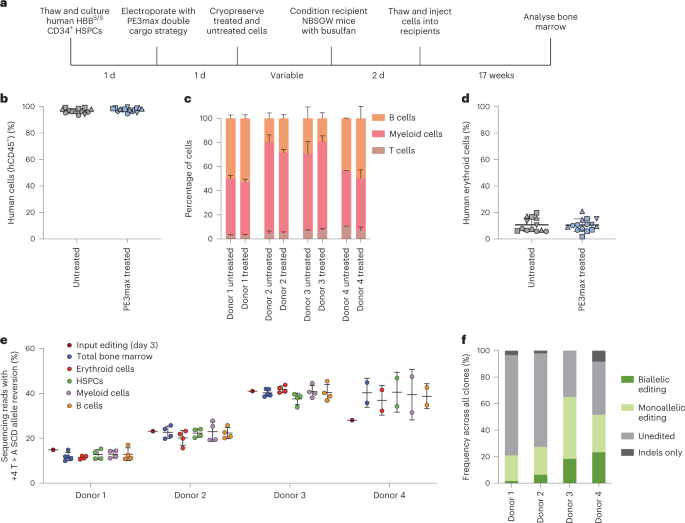
Figure 3 Engraftment of SCD Patient CD34+ HSPCs After Prime Editing and Transplantation into Immunodeficient Mice
EDITGENE, leveraging Prime Editing technology, has comprehensively upgraded its Bingo™ platform for site-specific mutagenesis. Drawing from extensive experiences in thousands of CRO projects, EDITGENE has optimize its technical approach, resulting in significantly higher success rates compared to conventional PE methods.
EDITGENE now offers precise and efficient gene point mutation cell line construction services to a broad spectrum of scientific research enterprises and communities.
Recent blogs :
1. [Literature Review] Multiplex Detection Strategy of Biosensors Based on CRISPR-Cas System
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com












![[Cutting-Edge News] Developments in Prime Editing - Prime Editing Offers New Approaches for Disease Treatment](/uploads/20241026/HQIYhofiABGLbU0C_6b1e7d3bcc99e579649fc9f8f24ceae0.jpg)

Comment (4)