[Star of the Month] Human Whole Genome CRISPR/Cas9 Knockout Library and Human RNA Binding Protein Library
CRISPR library screening

CRISPR library screening is a high-throughput gene screening method based on CRISPR/Cas9 technology. It involves creating a library with thousands of sgRNAs, which are cloned into lentiviral vectors and used to infect target cells at a low MOI (Multiplicity of Infection). This ensures that each cell is infected with a single sgRNA, allowing for precise functional screening of genes. In this article, we feature the top three best-selling libraries of the month, along with key research papers to help guide, give insights, and advance your scientific research.
I. Sequential genome-wide CRISPR-Cas9 screens identify genes regulating cell-surface expression of tetraspanins
Original Link: https://doi.org/10.1016/j.celrep.2023.112065
Four-transmembrane proteins belong to a superfamily of membrane proteins. They were first identified while searching for new cell surface antigens in mammalian cancer cells. Currently, 33 members of this four-transmembrane protein superfamily have been discovered in humans, named TSPAN1 through TSPAN33. These proteins mediate various biological processes, including immune responses and tumor development. However, the mechanisms that regulate their expression on the cell surface remain unclear. To identify the regulatory factors involved in the transport of four transmembrane proteins, researchers focused on TSPAN8.
MEC is a liver cancer cell line derived from a cholangiocarcinoma patient, and it expresses the highest levels of TSPAN8 at both the mRNA and protein levels among all liver cancer cell lines. To identify genes affecting TSPAN8 expression, the researchers performed a whole genome CRISPR-Cas9 knockout screen on MEC cells. The screening identified SPPL3, MGAT1, and MGAT2 as the most promising genes influencing TSPAN8 expression.
To validate whether these genes affect TSPAN8 expression, the researchers used CRISPR-Cas9 gene knockout and rescue experiments targeting MGAT1 and MGAT2. The results showed that knockout of MGAT1 and MGAT2 significantly reduced TSPAN8 expression. Moreover, re-expression of MGAT1 in the MGAT1-KO MEC cells fully restored TSPAN8 expression on the cell surface. These findings demonstrate that the MGAT1 and MGAT2 genes play crucial roles in regulating TSPAN8 expression.
In the article, the authors used the MEC whole-genome CRISPR-Cas9 knockout library to screen for key genes that regulate the expression of the four-transmembrane protein TSPAN8. They identified critical pathways that affect its expression, providing important insights into the mechanisms of tumor invasion.
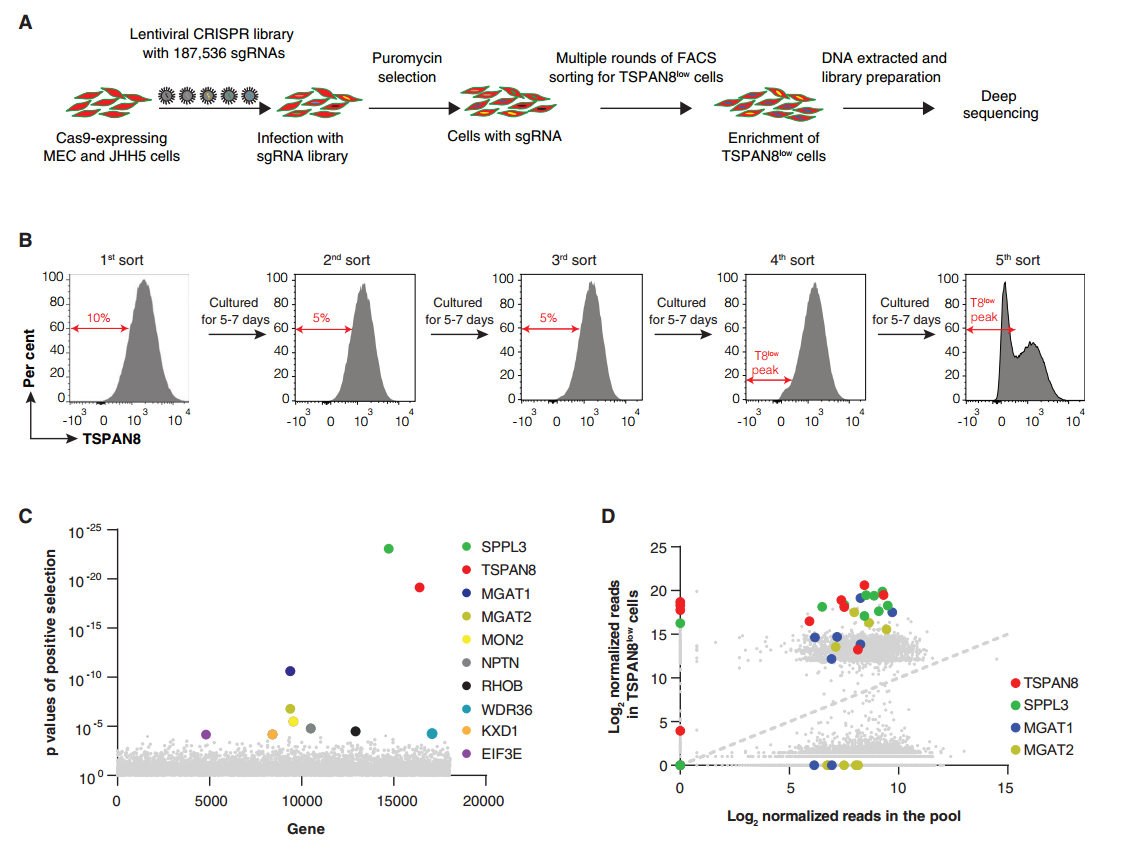
Figure 1: MEC Whole Genome CRISPR-Cas9 Knockout Screen Identifies Genes Regulating Cell Surface Expression of TSPAN8
II. Epigenomic reprogramming via HRP2-MINA dictates response to proteasome inhibitors in multiple myeloma with t(4;14) translocation
Original Link: https://doi.org/10.1172/JCI149526
Multiple myeloma (MM) is a genetically complex and heterogeneous hematologic malignancy, characterized by the monoclonal expansion of malignant plasma cells and a range of genetic abnormalities, including chromosomal translocations, deletions, duplications, and gene mutations. HRP2, also known as HDGFRP2, is a structural homolog of the hepatoma-derived growth factor-related protein family. In cancer cells, HRP2 promotes hepatocellular carcinoma growth by interacting with the RNA processing regulator IWS1. Nonetheless, the expression, biological function, and regulatory mechanisms of HRP2 in hematologic malignancies, especially in multiple myeloma, remain poorly understood.
In this study, a whole-genome CRISPR/Cas9 knockout screen using a library of 70,290 sgRNAs was employed to identify genes that regulate bortezomib sensitivity and resistance in multiple myeloma (MM). Through positive and negative selection, the researchers identified 28 genes associated with bortezomib sensitivity and 15 genes linked to resistance. HRP2 (HDGFRP2) was found to be the most differentially expressed gene in bortezomib-resistant MM cells. Further analysis revealed that HRP2 plays a key role in regulating H3K27me3 levels in genes involved in ER stress-related apoptosis, suggesting an epigenetic mechanism.
These findings highlight HRP2 as a critical gene influencing bortezomib sensitivity in MM, offering new insights into treatment mechanisms and potential therapeutic targets. This study utilizes human whole-genome CRISPR screening to advance our understanding of MM therapy and resistance.
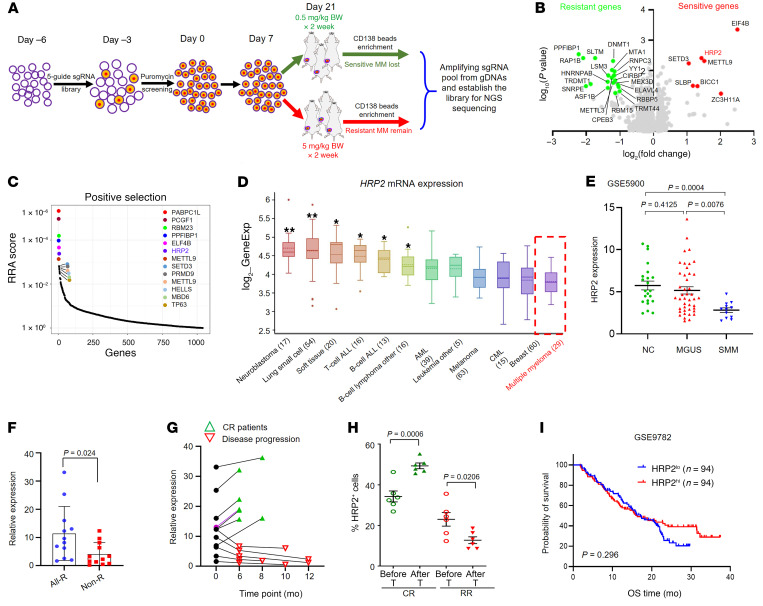
Figure 2: In vivo CRISPR library screening identifies HRP2 as a key negative regulator of bortezomib resistance.
III. CRISPR-Cas9 screening identifies INTS3 as an anti-apoptotic RNA-binding protein and therapeutic target for colorectal cancer
Original Link: https://doi.org/10.1016/j.isci.2024.109676
Colorectal cancer (CRC) is a malignant tumor originating from the epithelial cells of the colon or rectum, with high incidence and mortality rates. Studies have shown that certain RNA-binding proteins (RBPs) are critical for CRC cell survival, with key mechanisms including anti-apoptotic functions, tumor growth promotion, and RNA metabolism regulation. To identify CRC-related genes, the researchers conducted a series of studies.
The researchers used a CRISPR library containing 1,078 RBP-targeted sgRNAs to perform high-throughput screening in CRC cell lines. The goal was to identify key RBP genes that significantly affect cell survival or proliferation by measuring changes in sgRNA abundance. Through this CRISPR library screening, 27 RBPs associated with CRC cell survival were systematically identified, with the INTS3 gene showing the highest correlation.
To validate the role of INTS3, the researchers designed sgRNAs targeting the gene and found that its knockout significantly increased apoptosis in CRC cells and inhibited tumor growth both in vitro and in vivo. This further confirmed the critical role of INTS3 in cancer cell survival. By combining CRISPR-Cas9 knockout of INTS3 with transcriptome sequencing, the researchers discovered that INTS3 regulates the stability of anti-apoptotic gene mRNAs, such as TXNIP, CLU, and NR4A1, thereby enhancing CRC cell survival.
These findings provide an in-depth understanding of the functional roles of RBPs in CRC and offer new targets and strategies for cancer precision therapy. With its high throughput, specificity, and efficiency, CRISPR library screening technology has emerged as a cutting-edge tool for analyzing gene function networks and identifying key therapeutic targets, providing crucial support for precision medicine research.
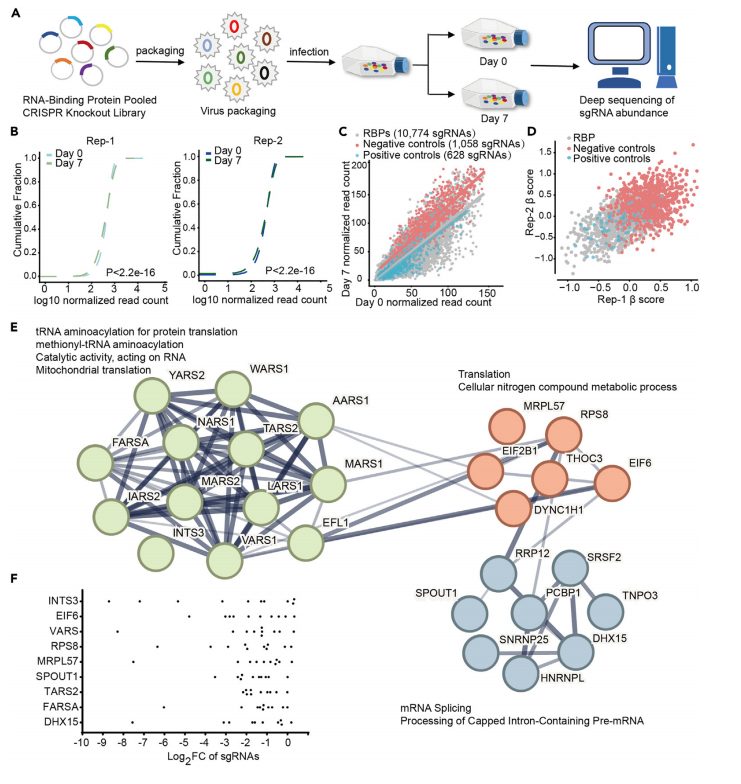
Figure 3: CRISPR-Cas9 Screening of RNA-Binding Proteins (RBPs) in Colorectal Cancer (CRC) Cells
EDITGENE has over a decade of expertise in gene editing and offers the most comprehensive series of efficient Cas9 stable cell lines, featuring the latest and most popular CRISPR sgRNA libraries . With a dedicated team of experts providing personalized service, you can start screening today!
Recent Blogs
1.Stuff Your Lab with Savings: Thanksgiving Sale on Engineered Cell Lines!
2.[Weekly News] Genome-Wide CRISPR Screening: Uncovering New Host Factors to Optimize Rotavirus Vaccine Production
3.[Literature Review] Unlocking CRISPR-Cas12a's Full Potential: Enhancing Trans-Cleavage with Variants
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com

















![[Literature Insight] From CRISPR Screening to FCD Mechanisms: Do Cilia Also Influence Nerves?](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)