[Literature Review] SATCAS: A CRISPR/Cas13a-based isothermal one-pot RNA detection platform
CRISPR/Cas13a

Rapid, cost-effective, and sensitive pathogen nucleic acid detection plays a crucial role in clinical diagnosis and infection management. Traditional CRISPR/Cas13a-based detection methods show great potential in molecular diagnostics; however, most require a transcription step, and the multi-step open operations are prone to contamination, which limits their widespread application. To overcome the limitations of existing technologies, Shenzhen Hospital of Peking University has collaborated with researchers from Peking University People's Hospital and Fudan University to propose an isothermal one-pot RNA detection platform—SATCAS—which provides a more effective method for pathogen and SNP detection in clinical diagnosis. The related research results, titled "SATCAS: A CRISPR/Cas13a-based simultaneous amplification and testing platform for one-pot RNA detection and SNPs distinguish in clinical diagnosis," were recently published in the journal Biosensors and Bioelectronics (IF: 10.7).
Original Article Link: https://doi.org/10.1016/j.bios.2024.116636
Serum hepatitis B virus (HBV), as a highly threatening infectious pathogen, is in urgent need of rapid and effective diagnostic methods for its suppression, and the detection of the pathogen RNA is of great significance. Serum hepatitis B virus (HBV) pregenomic RNA (pgRNA) can serve as an alternative biomarker reflecting HBV transcriptional activity. However, there are currently very few standardized methods available for quantifying serum HBV RNA in patients with hepatitis B disease. CRISPR-based molecular diagnostics demonstrate advantages such as good detection performance and low equipment costs, showing potential for point-of-care testing (POCT). However, there are very few CRISPR-Dx methods that can rapidly and specifically detect viable bacterial pathogens. To address this, the study combined simultaneous amplification and testing (SAT) with CRISPR/Cas13a to construct a one-pot isothermal nucleic acid detection platform called SATCAS, which can directly and quickly detect nearly single-copy RNA molecules in each reaction within 40 minutes.
1. Working Principle of SATCAS Platform
The SATCAS (Simultaneous Amplification and Testing platform based on Cas13a) platform works through enzyme cascade catalysis , which consists of two primers, reverse transcriptase, T7 RNA polymerase, CRISPR-Cas13a, and crRNA system. When the target RNA is present, the T7 promoter primer binds to the target RNA and generates an RNA/DNA hybrid double strand using reverse transcriptase. The RNase H activity of the reverse transcriptase specifically digests the hybridized RNA strand, leaving a single DNA strand containing T7 promoter. Next, a second primer binds to the DNA single strand and generates double-stranded DNA containing the T7 promoter by reverse transcription. After that, T7 RNA polymerase recognizes the T7 promoter sequence in the double-stranded DNA and continuously transcribes RNA from this template. Finally, the transcribed RNA is recognized by Cas13a, activating its cleavage activity, cleaving the fluorescent reporter, and emitting a detectable fluorescent signal.
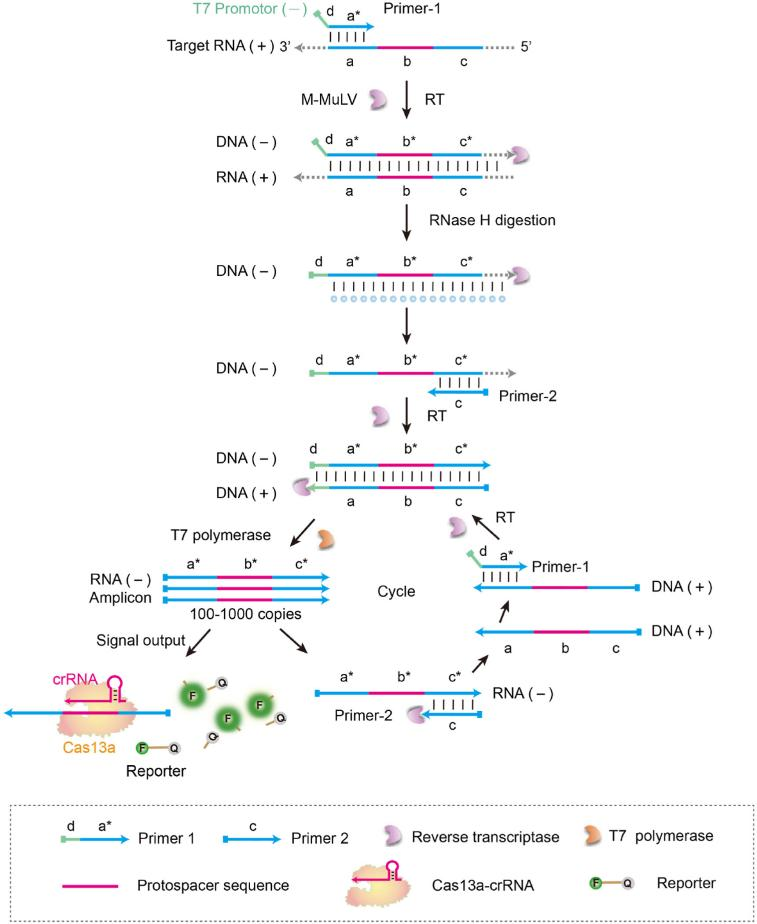
Fig.1 Schematic diagram of SATCAS for rapid and visual RNA detection
2. Exploration of One-Pot Detection and SATCAS Feasibility Verification
To avoid contamination during the two-step process, the researchers attempted to integrate the SAT reaction with the Cas13a-mediated cleavage reaction system, but the results were unsatisfactory. After multiple optimizations, they finally decided to separate the Cas13a system from the SAT amplification system, placing them at the bottom and lid of the tube, respectively. The detection procedure was as follows: After 30 minutes of SAT amplification, the Cas13a cleavage system was mixed with the SAT product through oscillation, followed by a 5-10 minute Cas13a cleavage reaction at 37°C. Finally, the fluorescence results were observed visually under blue light.
In addition, to invest the feasibility of SATCAS, the researchers used SARS-CoV-2 fragments as DNA templates for agarose gel electrophoresis and real-time fluorescence detection. The results showed that only the complete CRISPR/Cas13a system in the presence of the target produced a significant fluorescent signal, demonstrating the feasibility of SATCAS assay. By using the HBV-specific SATCAS assay to detect other common viral RNAs, it was found that SATCAS had almost no fluorescent response to interfering viral RNAs, demonstrating the high specificity of SATCAS.
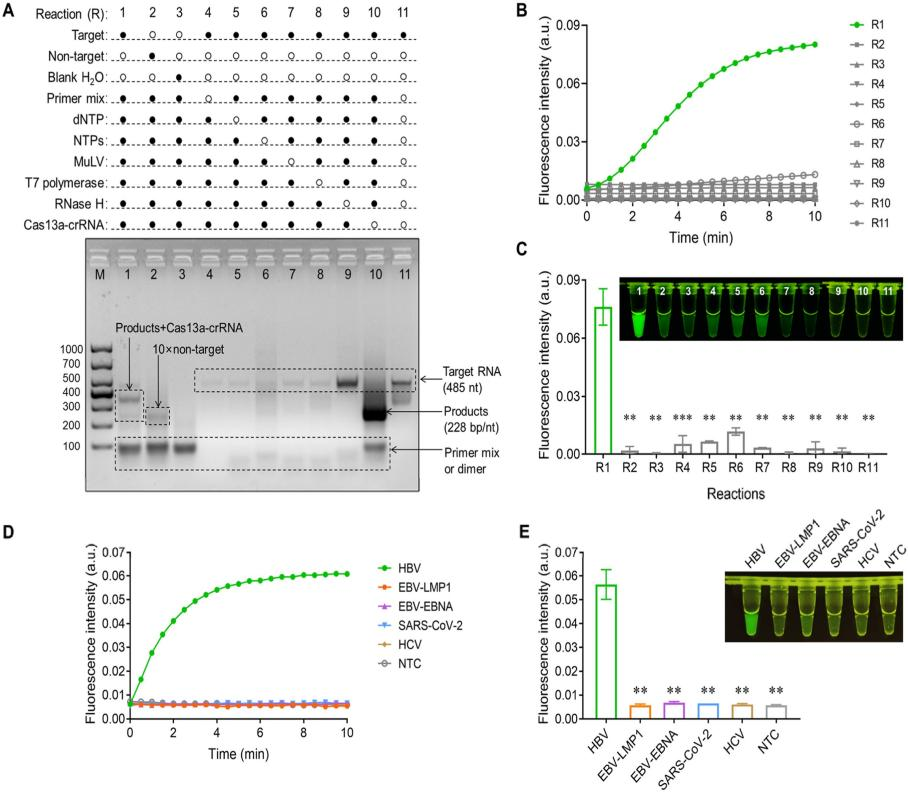
Fig.2 Feasibility investigation of SATCAS
3. Detection of Live Bacteria
RNA detection is the sole method for detecting viable pathogens. To validate the ability of SATCAS to distinguish between live and dead bacteria, the researchers prepared specimens containing various proportions of live and dead Escherichia coli (ATCC 25922) for detection, the ratio of viable to total bacteria ranged from 0% to 100%. Initially, the samples were analyzed using standard plate count and qRT-PCR methods, but no significant differences were found. This was attributed to the presence of the target DNA in both live and dead bacteria. Subsequently, total RNA extracted from these samples was analyzed using the SATCAS method, revealing an increase in fluorescence intensity with increasing proportions of viable bacteria. This indicates that the S ATCAS method was able to quantify the number of live bacteria.
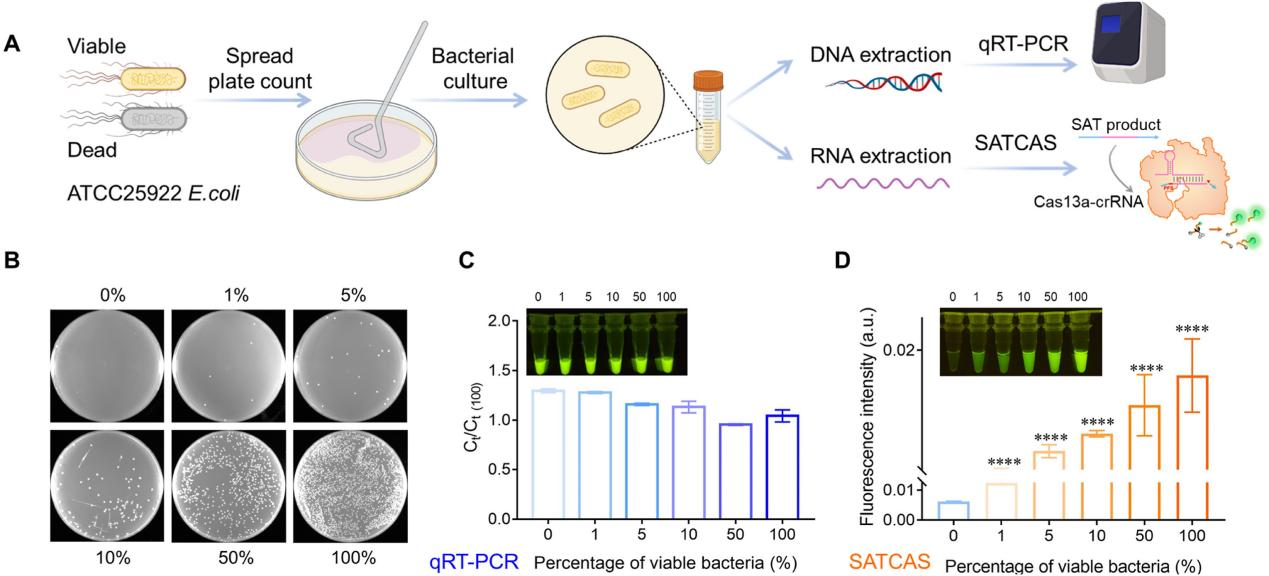
Fig3. Quantification of viable pathogenic bacteria
4. crRNA Screening and Sensitivity Testing
To achieve better sensitivity of the SATCAS assay, the researchers optimized the crRNA screening. They designed crRNA sequences targeting the SARS-CoV-2 amplification fragments and tested the corresponding real-time fluorescence values and fluorescence intensity visualization results. The results indicated that crRNA5 performed best. According to the optimal crRNA5, the researchers detected SARS-CoV-2 RNA at different concentration gradients to determine the sensitivity of the SATCAS method. The test results showed that SATCAS could consistently detect 10^10nM (0.1 aM) of target RNA in a 40 μL reaction system, indicating a limit of detection (LOD) of 2.41 RNA copies/40 μL, comparable to qRT-PCR and LAMP, and superior to some previously reported CRISPR-based methods . Similarly, the researchers also screened crRNAs of HBV (hepatitis B virus) and determined the optimal crRNA2 and its detection sensitivity.
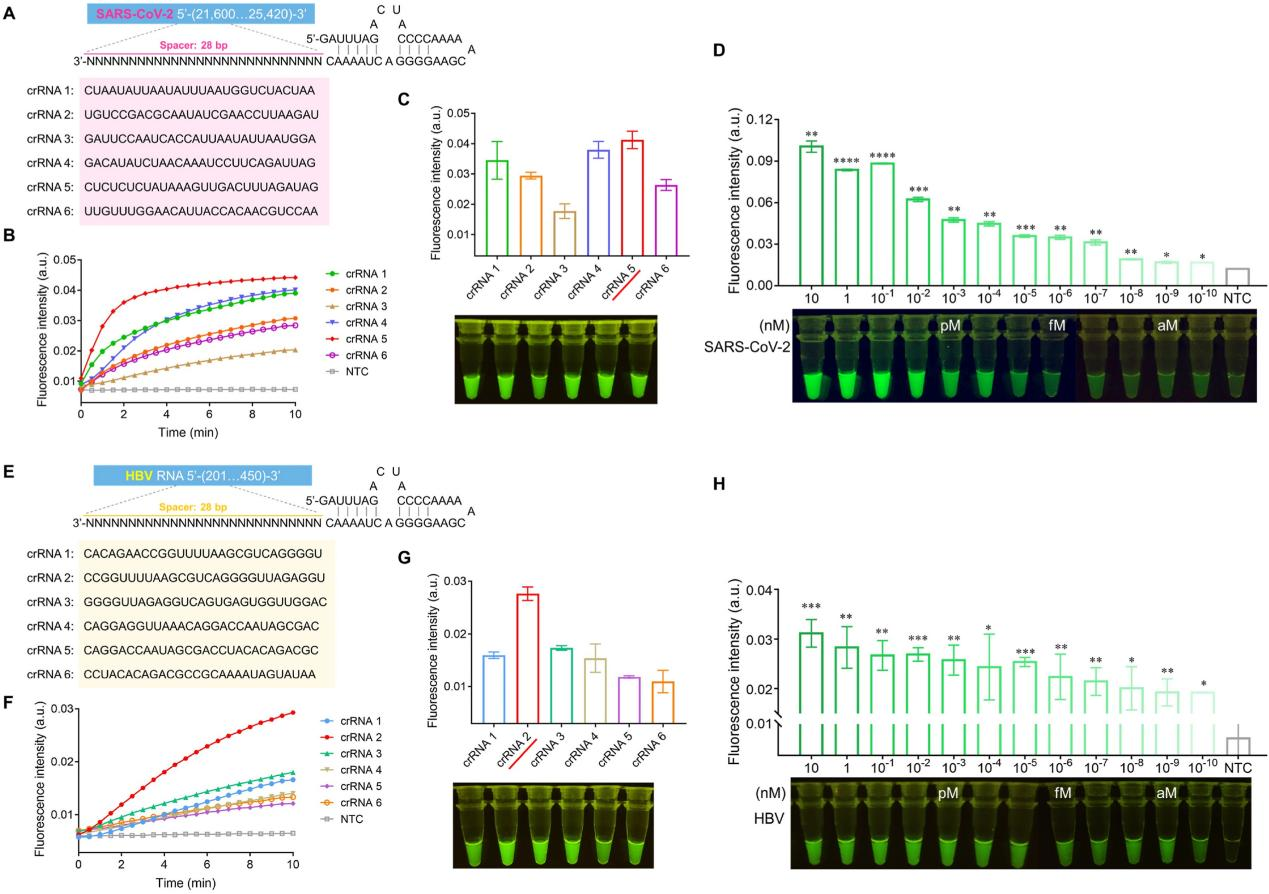
Fig4. Optimization of crRNAs and exploration of detection sensitivity of SARS-CoV-2 and HBV
5. Specificity Evaluation of SATCAS
LwaCas13a exhibited a remarkable 70% single-nucleotide mutation identification at position 14 of its recognized region. However, it generally demonstrates tolerance to single nucleotide mismatches, posing a significant challenge in distinguishing single nucleotide polymorphisms (SNPs). To evaluate the specificity of SATCAS in detecting the HBV drug resistance mutation site L180M, the researchers designed a series of CRISPR RNA (crRNA) by introducing the L180M mutation site at the 14th position of the crRNA recognition region, they assessed the detection efficiency of these crRNAs against both wild-type (WT) and L180M mutant targets. Comparative analysis of endpoint fluorescence values and visualized fluorescence intensities revealed that certain crRNAs exhibited significantly distinct detection intensities between WT and mutant tar gets, indicating a high specificity ratio . To further validate SATCAS's ability to identify single nucleotide mutations in detecting HBV drug resistance-related tyrosine-methionine-aspartic acid (YMDD) variants, researchers found that these variants may lead to the virus developing resistance to lamivudine, resulting in viral rebound, replication, and even disease progression.
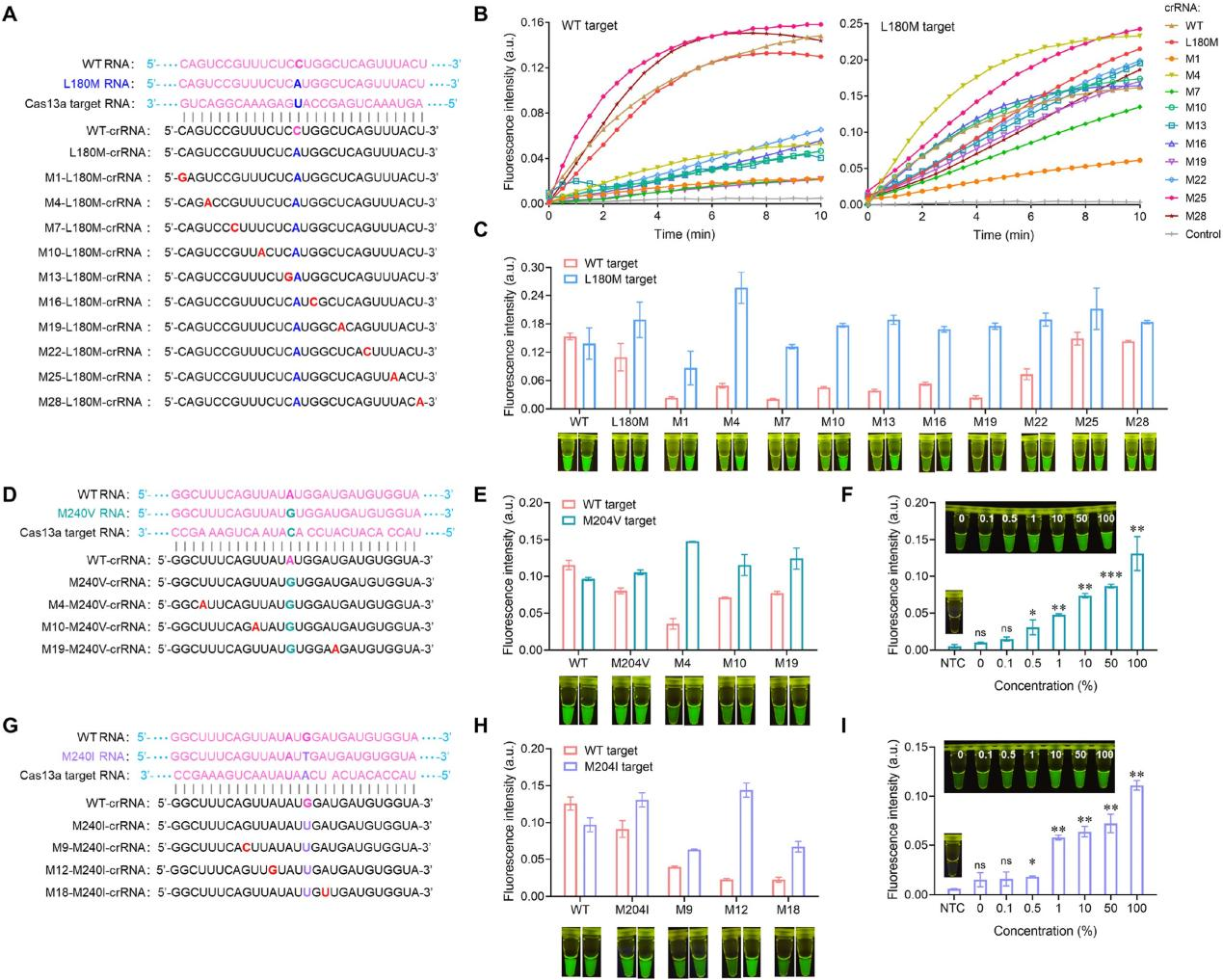
Fig5. Specificity investigation of SATCAS and its application in the identification of HBV
6. Evaluation of SATCAS Application Capabilities
To evaluate the application capabilities of the SATCAS method in detecting drug resistance mutations, the researchers conducted detection and validation experiments on HBV YMDD variants, different crRNA mutation templates, and mutation templates at various concentrations. All results demonstrated that the SATCAS method could accurately and sensitively detect drug resistance mutations. Additionally, the researchers evaluated the application of the SATCAS method in detecting HBV and EBV viruses. They collected blood samples from HBV-infected patients and performed parallel detections using the SATCAS method and qRT-PCR. Comparative analysis revealed a high degree of consistency between the results of the two methods, with SATCAS exhibiting 100% sensitivity, 92.86% specificity, and 97.06% accuracy. Correlation analysis showed a strong or moderate correlation between the two methods. Comparisons with clinical samples further supported the high sensitivity, specificity, and robustness of the SATCAS method in viral infection diagnosis and validated its application potential in RNA detectio n .
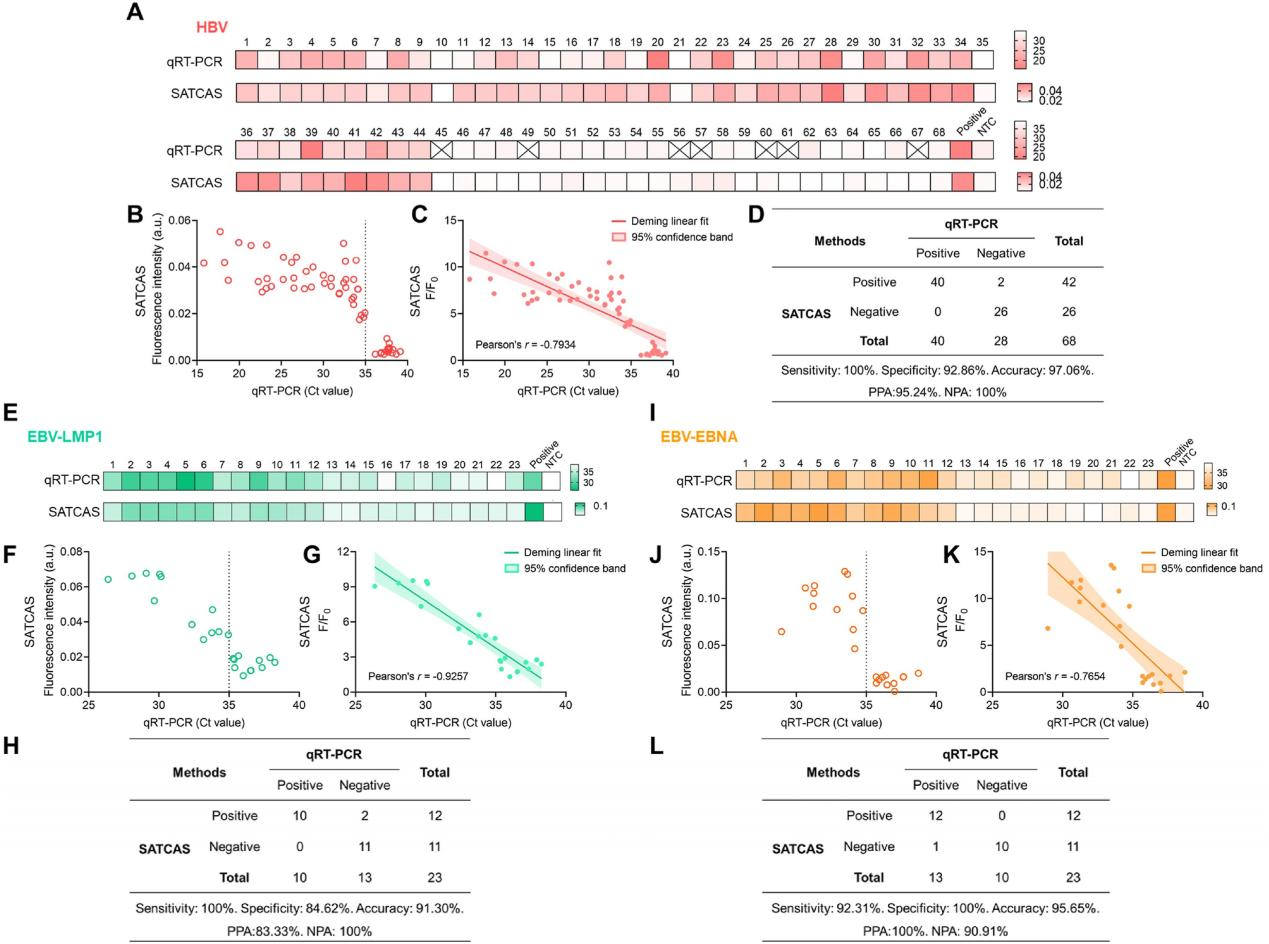
Fig.6 Robust performance of the SATCAS assay for detection of HBV and EBV in clinical specimens
In summary, according to the need to detect and differentiate viral mutations associated with drug resistance, the researchers developed an isothermal one-pot RNA detection platform-SATCAS-by combining SAT with CRISPR/Cas13a . SATCAS has demonstrated its exceptional sensitivity, robustness against interference, easy-operation, and visualization for nucleic acid detection, making it suitable for live pathogen detection and clinical diagnosis of viral infections. Although SATCAS still requires an additional step of RNA extraction from clinical samples, it shows great potential as a routine quantitative RNA detection method.
EDITGENE has been specialized in CRISPR/Cas Gene Editing for more than 10 years, we provide our service based on protein purification platform, we provide innovative one-step purification of Cas enzyme , realizes significant improvement in purity, activity and recycle rate, with performance far exceeding that of similar products in the international market, and sensitivity up to amol level . Our innovative service is widely used in the fields of CRISPR research and development, gene and disease detection, and food safety testing. We also have thermostatic rapid amplification kits and CRISPR detection kits in stock, with low equipment requirements and short reaction time, which can be delivered immediately upon placing an order!
Recent Blogs :
2. [Literature Review] CRISPR KO Screening Reveals Key Genes Responsible for Chemotherapy Resistance
Follow us on social media
Contact us
+ 833-226-3234 (USA Toll-free)
+1-224-345-1927 (USA)
info@editxor.com















Comment (4)