[Literature Review] A Novel Mechanism of Cisplatin Resistance in Osteosarcoma: CRISPR Screening Identifies Key Regulators in Organoid Models
CRISPR screening

Osteosarcoma is the most common primary malignant bone tumor in adolescents, and cisplatin-based chemotherapy remains a cornerstone of clinical treatment. However, the development of cisplatin resistance often leads to disease relapse or metastasis, significantly compromising treatment outcomes. To date, the molecular mechanisms underlying this resistance remain largely unclear, and effective models to study the resistance process are lacking.
In response to this challenge, a research team from the Second Xiangya Hospital developed a patient-derived osteosarcoma organoid model and induced cisplatin resistance within it. By integrating this model with CRISPR screening technology, they successfully identified key regulators driving cisplatin resistance. The team further explored the mechanisms of these critical factors, providing valuable insights for the development of targeted therapies for patients with resistant osteosarcoma.
Their findings, titled “A Dual Approach with Organoid and CRISPR Screening Reveals ERCC6 as a Determinant of Cisplatin Resistance in Osteosarcoma,” were recently published in Advanced Science (Weinheim, Baden-Wurttemberg, Germany; Impact Factor: 14.3).
Original Link: https://doi.org/10.1002/advs.202500632
Spotlight
1. Innovative Model: This study is the first to induce cisplatin resistance in osteosarcoma organoids (OSO). Single-cell RNA sequencing revealed heterogeneity among patient-derived organoids, establishing a clinically relevant resistance model.
2. Methodological Advancement: A dual approach combining drug-induced organoid resistance and CRISPR screening precisely identified the resistance gene ERCC6.
3. Mechanistic Insight: The study uncovers a novel mechanism whereby ERCC6 and HNRNPM regulate alternative splicing of BAX pre-mRNA and the PI3K/AKT pathway, offering new therapeutic targets to overcome cisplatin resistance.
Compared with traditional cell line models, organoid models better recapitulate the cellular composition and heterogeneity of tumors, exhibiting higher clinical relevance and biological fidelity.
To investigate cisplatin resistance in depth, the researchers established organoid models derived from osteosarcoma patient tumor samples. These models were validated using a series of techniques including single-cell RNA sequencing (scRNA-seq), RNA sequencing (RNA-seq), and histopathological analyses.
Gene expression profiling demonstrated that the organoids closely resembled the original tumors. Heatmaps of the top 25 upregulated and downregulated genes in tumor tissue compared to normal tissue showed consistent expression patterns in the organoids (Figure 1E). UMAP visualization and bar charts revealed the presence of all major cell types except vascular-associated cells within the organoids (Figures 1F and 1G).
Furthermore, immunohistochemistry (IHC) and immunofluorescence (IF) staining confirmed a high concordance in the expression of key markers—including CD99, VIM, CD68, KI67, and SATB2—between the organoids and the tumor tissues.
Collectively, these results indicate that the organoid model faithfully mirrors the gene expression, cellular composition, and histological features of the primary tumor, supporting its clinical relevance.
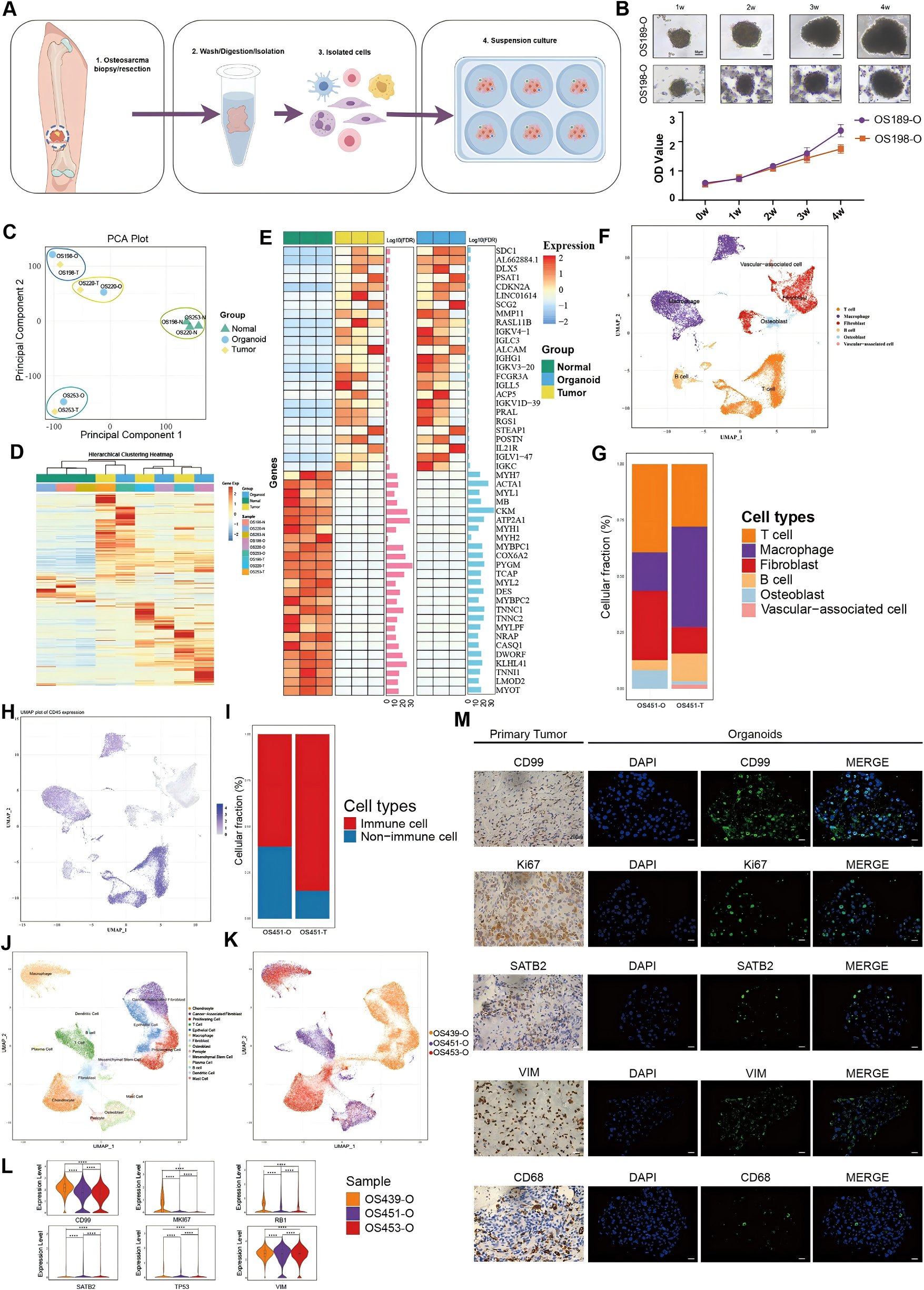
Figure 1. Establishment and characterization of OSO
After successfully validating the high reliability and clinical relevance of the organoid models, the researchers further simulated the clinical drug resistance process to investigate the mechanism underlying cisplatin resistance.
Organoids derived from three patients were divided into two groups: one continuously exposed to cisplatin, and the other left untreated as a control. RNA sequencing analysis revealed significant differences in gene expression profiles between resistant and non-resistant organoids. Differential expression analysis identified genes that were significantly upregulated or downregulated in the resistant organoids (Figure 2E).
Subsequently, a genome-wide CRISPR screen was conducted under cisplatin treatment. By integrating gene enrichment analysis with CRISPR screening results, ERCC6 was identified as the sole gene upregulated in the resistant organoids. Together with immunohistochemistry (IHC) scoring (Figure 2K), these findings suggest a correlation between ERCC6 expression and chemoresistance.
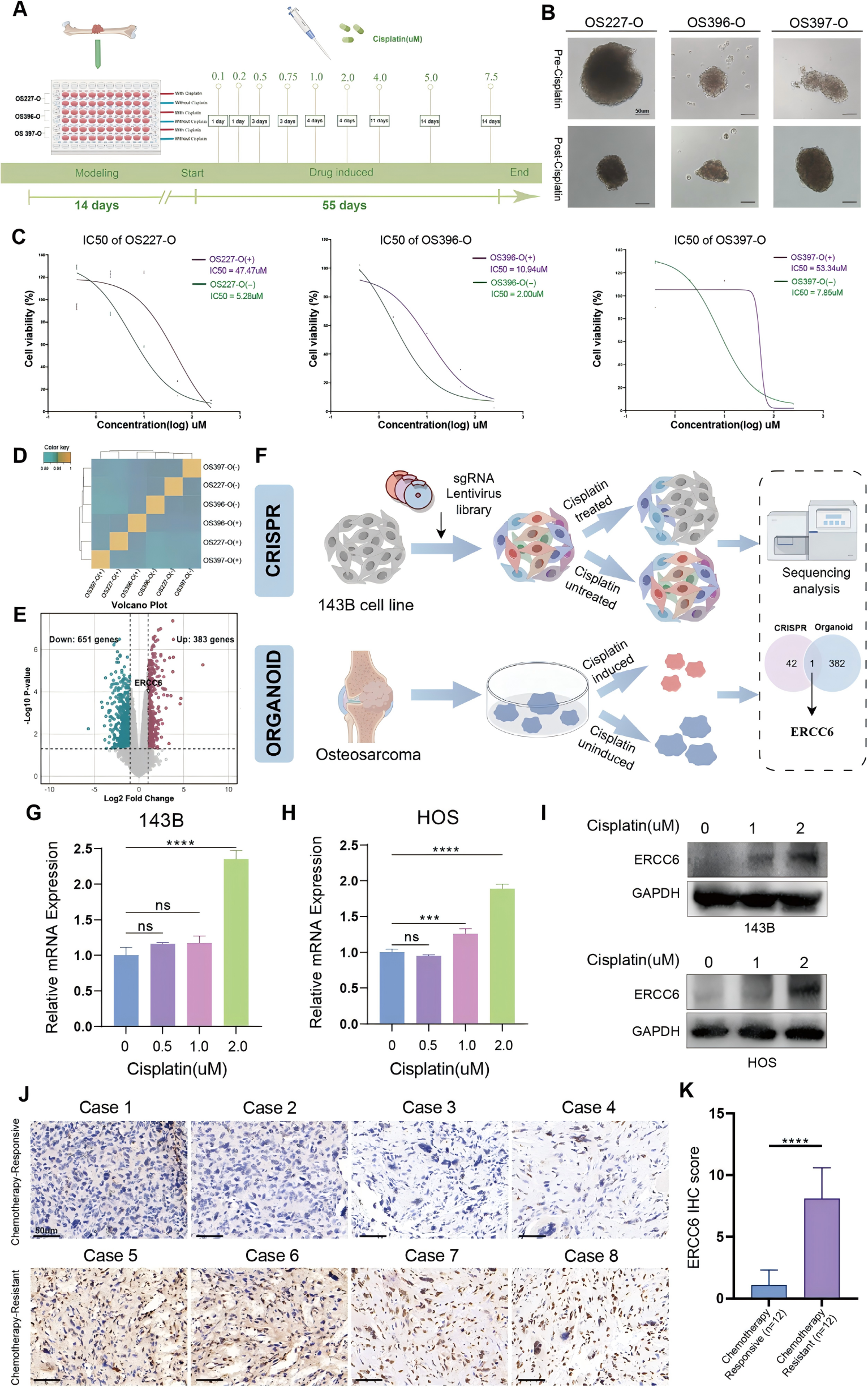
Figure 2. Identification of key genes associated with cisplatin resistance
To investigate the role of ERCC6 in cisplatin resistance and verify its functional relevance, both in vitro and in vivo experiments were conducted.
First, researchers used the CRISPR/Cas9 system to knockout ERCC6 in 143B and HOS osteosarcoma cell lines, and successful knockout was confirmed via Western blot analysis. Following ERCC6 knockout, the IC50 values of 143B and HOS cells decreased significantly. Under cisplatin treatment, the proliferation rate and colony formation ability of both cell lines were markedly reduced, while apoptosis rates significantly increased.
To further validate these findings in vivo, a subcutaneous xenograft model was established by injecting ERCC6-knockout or control 143B cells into mice. Mice in the treatment group received intraperitoneal injections of cisplatin (2 mg/kg) every three days, while the control group received saline. Tumor volume and weight were monitored throughout the experiment.
The results showed that ERCC6 knockout significantly suppressed tumor growth and reduced tumor weight in the cisplatin-treated group. Collectively, these findings confirm that ERCC6 knockout significantly enhances the sensitivity of osteosarcoma cells to cisplatin.
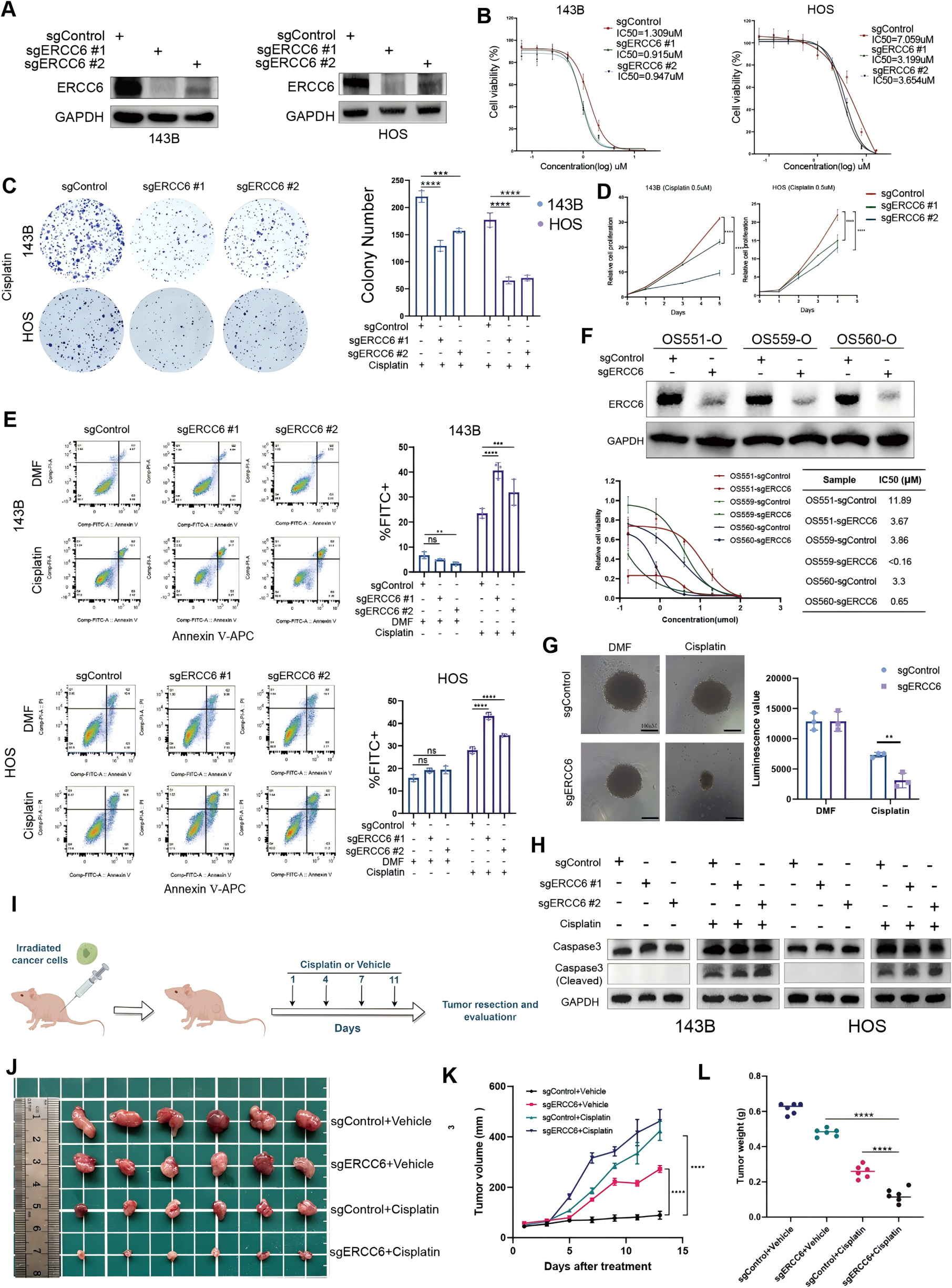
Figure 3. Functional validation of ERCC6 in cisplatin sensitivity
To further elucidate the mechanism by which ERCC6 contributes to cisplatin resistance, the researchers conducted a comprehensive study using protein interaction assays, RNA sequencing, and molecular docking analyses. These approaches were used to explore the interaction between ERCC6 and HNRNPM, as well as their roles in regulating the PI3K/AKT signaling pathway and alternative splicing of BAX.
The study first examined the relationship between ERCC6 and the PI3K/AKT pathway. Gene expression profiling revealed a significant downregulation of PI3K/AKT pathway-related genes in ERCC6-knockout cells. This was further validated by Western blot analysis, which showed a marked decrease in phosphorylated AKT (p-AKT) levels upon ERCC6 knockout, while total AKT levels remained unchanged. These findings suggest a critical role for ERCC6 in modulating the PI3K/AKT pathway.
The interaction between ERCC6 and HNRNPM was then investigated in detail. Co-immunoprecipitation (Co-IP) and mass spectrometry analyses identified HNRNPM as a binding partner of ERCC6. A GST pull-down assay further confirmed that the nucleotide-binding domain (NBD; amino acids 1–510, designated D1) of ERCC6 is essential for its interaction with HNRNPM (Figure 4J). Additionally, immunofluorescence co-localization experiments clearly demonstrated the subcellular co-localization of ERCC6 and HNRNPM.
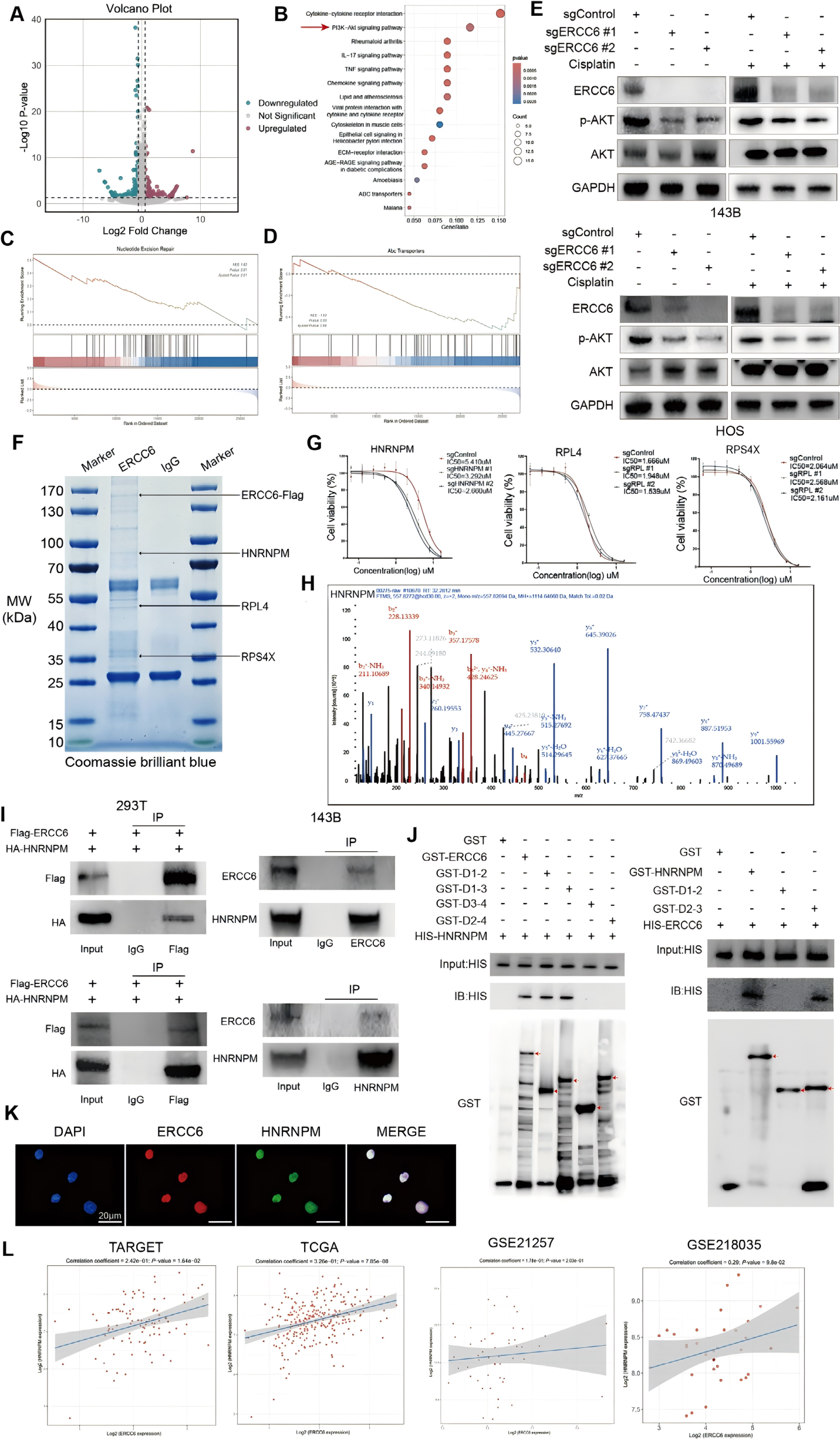
Figure 4. ERCC6 Regulates the AKT Pathway and Interacts with Binding Partner HNRNPM
To further elucidate the downstream effects of the ERCC6–HNRNPM interaction, RNA sequencing was employed to analyze alternative splicing events in ERCC6- and HNRNPM-knockout cells. The results revealed that knockout of either ERCC6 or HNRNPM significantly affected the alternative splicing of BAX, particularly promoting exon 2 skipping events.
As shown in Figure 5, qRT-PCR and agarose gel electrophoresis demonstrated a marked increase in the expression of the full-length BAX transcript following the knockout of ERCC6 and HNRNPM. Moreover, Western blot analysis confirmed that BAX protein levels were significantly elevated, while BCL2 protein levels remained unchanged.
These findings highlight the crucial roles of ERCC6 and HNRNPM in regulating BAX alternative splicing and protein expression. They also uncover a dual mechanism through which ERCC6 promotes apoptosis and counteracts cisplatin resistance by suppressing the PI3K/AKT signaling pathway and modulating the alternative splicing of the pro-apoptotic gene BAX.
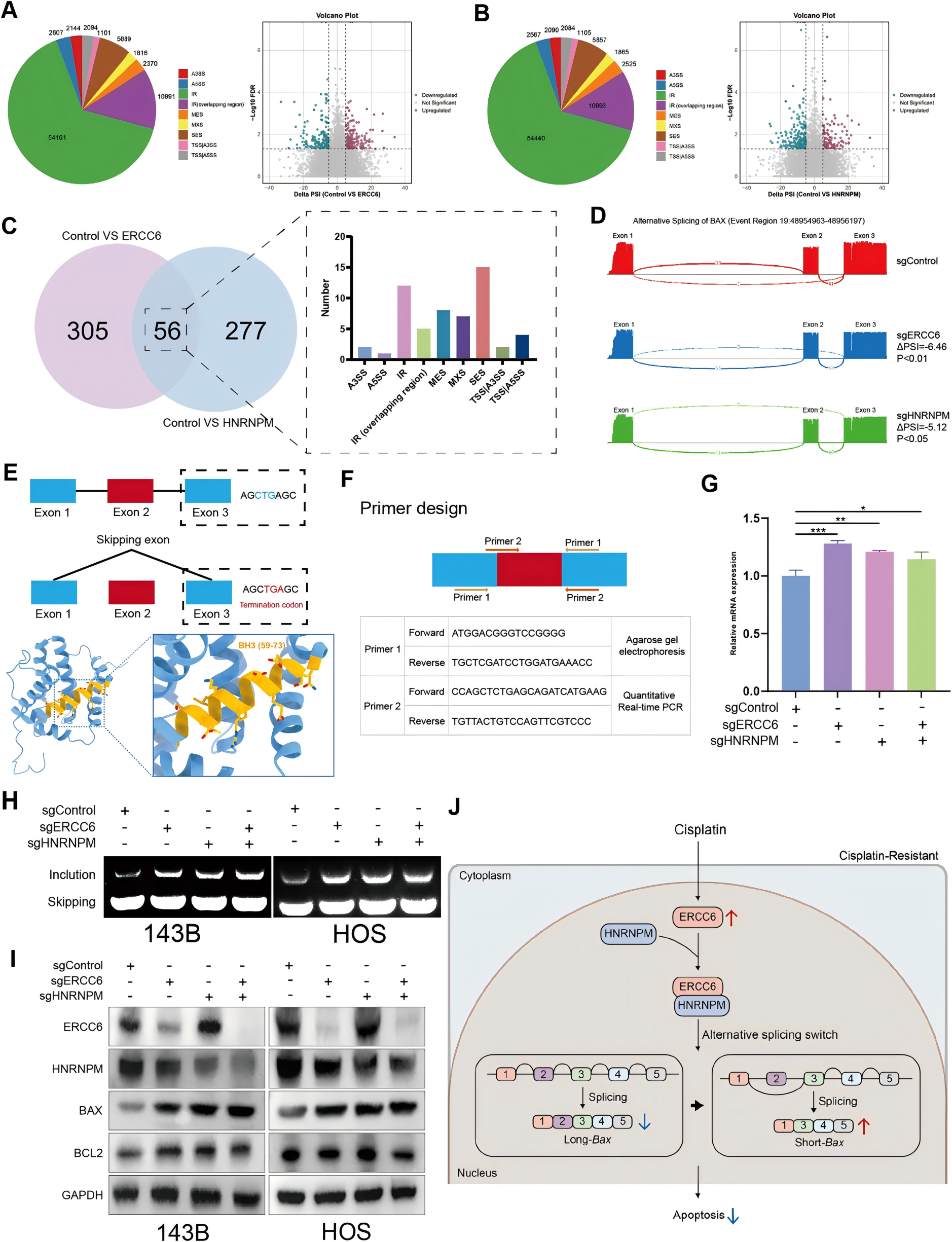
Figure 5. Regulation of alternative splicing by ERCC6 and HNRNPM
This study uncovers key molecular mechanisms underlying chemotherapy resistance and highlights ERCC6 and HNRNPM as promising therapeutic targets. The findings offer new insights into overcoming drug resistance in osteosarcoma and set the stage for future research.
Moving forward, the use of organoid models derived from other disease contexts may further advance our understanding of resistance mechanisms, paving the way for personalized drug screening and precision oncology.















![[Customer Publication] Methylation “Fingerprint” Detection May Transform Early Cancer Screening](/uploads/20250527/bL2GJjteMDvzmZys_53c82bdd67704fe0e159246934f924ee.png)

Comment (4)